Proper Division Plane Orientation and Mitotic Progression Together Allow Normal Growth of Maize
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Keystone Review Module B BIO.B.1.1 – Describe the Three Stages of the Cell Cycle: Interphase, Nuclear Division, Cytokinesis
Keystone Review Module B BIO.B.1.1 – Describe the three stages of the cell cycle: interphase, nuclear division, cytokinesis. ● Describe the events that occur during the cell cycle: interphase, nuclear division, and cytokinesis. ● Compare the processes and outcomes of mitotic and meiotic nuclear division. Which statement BEST describes the phase of the cell cycle shown? A. The cell is in prophase of mitosis because the number of chromosomes has doubled. B. The cell is in prophase I of meiosis because of the number of chromosomes has doubled. C. The cell is in telophase of mitosis because the cell is separating and contains two copies of each chromosome. D. The cell is in telophase of meiosis because the cell is separating and contains two copies of each chromosome. Answer - C A. Incorrect - The cell is not in prophase. This is obvious as the cell contains two nuclei, a condition which only occurs in telophase. B. Incorrect - The cell is not in prophase. This is obvious as the cell contains two nuclei, a condition which only occurs in telophase. C. Correct - The cell is in telophase, which can be seen from the two nuclei. Only telophase of mitosis includes two copies of each chromosome. D. Incorrect - The cell is in telophase, but in meiosis each cell contains only 1 copy of each chromosome. Mitosis and meiosis are processes by which animal and plant cells divide. Which statement best describes a difference between mitosis and meiosis? A. Meiosis is a multi-step process. B. Mitosis occurs only in eukaryotic cells. C. Meiosis is used in the repair of an organism. -

Mitosis Vs. Meiosis
Mitosis vs. Meiosis In order for organisms to continue growing and/or replace cells that are dead or beyond repair, cells must replicate, or make identical copies of themselves. In order to do this and maintain the proper number of chromosomes, the cells of eukaryotes must undergo mitosis to divide up their DNA. The dividing of the DNA ensures that both the “old” cell (parent cell) and the “new” cells (daughter cells) have the same genetic makeup and both will be diploid, or containing the same number of chromosomes as the parent cell. For reproduction of an organism to occur, the original parent cell will undergo Meiosis to create 4 new daughter cells with a slightly different genetic makeup in order to ensure genetic diversity when fertilization occurs. The four daughter cells will be haploid, or containing half the number of chromosomes as the parent cell. The difference between the two processes is that mitosis occurs in non-reproductive cells, or somatic cells, and meiosis occurs in the cells that participate in sexual reproduction, or germ cells. The Somatic Cell Cycle (Mitosis) The somatic cell cycle consists of 3 phases: interphase, m phase, and cytokinesis. 1. Interphase: Interphase is considered the non-dividing phase of the cell cycle. It is not a part of the actual process of mitosis, but it readies the cell for mitosis. It is made up of 3 sub-phases: • G1 Phase: In G1, the cell is growing. In most organisms, the majority of the cell’s life span is spent in G1. • S Phase: In each human somatic cell, there are 23 pairs of chromosomes; one chromosome comes from the mother and one comes from the father. -

The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression
International Journal of Molecular Sciences Review The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression Tingting Zou and Zhenghong Lin * School of Life Sciences, Chongqing University, Chongqing 401331, China; [email protected] * Correspondence: [email protected] Abstract: The cell cycle is a collection of events by which cellular components such as genetic materials and cytoplasmic components are accurately divided into two daughter cells. The cell cycle transition is primarily driven by the activation of cyclin-dependent kinases (CDKs), which activities are regulated by the ubiquitin-mediated proteolysis of key regulators such as cyclins, CDK inhibitors (CKIs), other kinases and phosphatases. Thus, the ubiquitin-proteasome system (UPS) plays a pivotal role in the regulation of the cell cycle progression via recognition, interaction, and ubiquitination or deubiquitination of key proteins. The illegitimate degradation of tumor suppressor or abnormally high accumulation of oncoproteins often results in deregulation of cell proliferation, genomic instability, and cancer occurrence. In this review, we demonstrate the diversity and complexity of the regulation of UPS machinery of the cell cycle. A profound understanding of the ubiquitination machinery will provide new insights into the regulation of the cell cycle transition, cancer treatment, and the development of anti-cancer drugs. Keywords: cell cycle regulation; CDKs; cyclins; CKIs; UPS; E3 ubiquitin ligases; Deubiquitinases (DUBs) Citation: Zou, T.; Lin, Z. The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression. 1. Introduction Int. J. Mol. Sci. 2021, 22, 5754. https://doi.org/10.3390/ijms22115754 The cell cycle is a ubiquitous, complex, and highly regulated process that is involved in the sequential events during which a cell duplicates its genetic materials, grows, and di- Academic Editors: Kwang-Hyun Bae vides into two daughter cells. -

Cell Division- Ch 5
Cell Division- Mitosis and Meiosis When do cells divide? Cell size . One of most important factors affecting size of the cell is size of cell membrane . Cell must remain relatively small to survive (why?) – Cell membrane has to be big enough to take in nutrients and eliminate wastes – As cells get bigger, the volume increases faster than the surface area – Small cells have a larger surface area to volume ratio than larger cells to help with nutrient intake and waste elimination . When a cell reaches its max size, the nucleus starts cell division: called MITOSIS or MEIOSIS Mitosis . General Information – Occurs in somatic (body) cells ONLY!! – Nickname: called “normal” cell division – Produces somatic cells only . Background Info – Starts with somatic cell in DIPLOID (2n) state . Cell contains homologous chromosomes- chromosomes that control the same traits but not necessarily in the same way . 1 set from mom and 1 set from dad – Ends in diploid (2n) state as SOMATIC cells – Goes through one set of divisions – Start with 1 cell and end with 2 cells Mitosis (cont.) . Accounts for three essential life processes – Growth . Result of cell producing new cells . Develop specialized shapes/functions in a process called differentiation . Rate of cell division controlled by GH (Growth Hormone) which is produced in the pituitary gland . Ex. Nerve cell, intestinal cell, etc. – Repair . Cell regenerates at the site of injury . Ex. Skin (replaced every 28 days), blood vessels, bone Mitosis (cont.) – Reproduction . Asexual – Offspring produced by only one parent – Produce offspring that are genetically identical – MITOSIS – Ex. Bacteria, fungi, certain plants and animals . -

List, Describe, Diagram, and Identify the Stages of Meiosis
Meiosis and Sexual Life Cycles Objective # 1 In this topic we will examine a second type of cell division used by eukaryotic List, describe, diagram, and cells: meiosis. identify the stages of meiosis. In addition, we will see how the 2 types of eukaryotic cell division, mitosis and meiosis, are involved in transmitting genetic information from one generation to the next during eukaryotic life cycles. 1 2 Objective 1 Objective 1 Overview of meiosis in a cell where 2N = 6 Only diploid cells can divide by meiosis. We will examine the stages of meiosis in DNA duplication a diploid cell where 2N = 6 during interphase Meiosis involves 2 consecutive cell divisions. Since the DNA is duplicated Meiosis II only prior to the first division, the final result is 4 haploid cells: Meiosis I 3 After meiosis I the cells are haploid. 4 Objective 1, Stages of Meiosis Objective 1, Stages of Meiosis Prophase I: ¾ Chromosomes condense. Because of replication during interphase, each chromosome consists of 2 sister chromatids joined by a centromere. ¾ Synapsis – the 2 members of each homologous pair of chromosomes line up side-by-side to form a tetrad consisting of 4 chromatids: 5 6 1 Objective 1, Stages of Meiosis Objective 1, Stages of Meiosis Prophase I: ¾ During synapsis, sometimes there is an exchange of homologous parts between non-sister chromatids. This exchange is called crossing over. 7 8 Objective 1, Stages of Meiosis Objective 1, Stages of Meiosis (2N=6) Prophase I: ¾ the spindle apparatus begins to form. ¾ the nuclear membrane breaks down: Prophase I 9 10 Objective 1, Stages of Meiosis Objective 1, 4 Possible Metaphase I Arrangements: Metaphase I: ¾ chromosomes line up along the equatorial plate in pairs, i.e. -

The Cell Cycle Coloring Worksheet
Name: Date: Period: The Cell Cycle Coloring Worksheet Label the diagram below with the following labels: Anaphase Interphase Mitosis Cell division (M Phase) Interphase Prophase Cytokinesis Interphase S-DNA replication G1 – cell grows Metaphase Telophase G2 – prepares for mitosis Then on the diagram, lightly color the G1 phase BLUE, the S phase YELLOW, the G2 phase RED, and the stages of mitosis ORANGE. Color the arrows indicating all of the interphases in GREEN. Color the part of the arrow indicating mitosis PURPLE and the part of the arrow indicating cytokinesis YELLOW. M-PHASE YELLOW: GREEN: CYTOKINESIS INTERPHASE PURPLE: TELOPHASE MITOSIS ANAPHASE ORANGE METAPHASE BLUE: G1: GROWS PROPHASE PURPLE MITOSIS RED:G2: PREPARES GREEN: FOR MITOSIS INTERPHASE YELLOW: S PHASE: DNA REPLICATION GREEN: INTERPHASE Use the diagram and your notes to answer the following questions. 1. What is a series of events that cells go through as they grow and divide? CELL CYCLE 2. What is the longest stage of the cell cycle called? INTERPHASE 3. During what stage does the G1, S, and G2 phases happen? INTERPHASE 4. During what phase of the cell cycle does mitosis and cytokinesis occur? M-PHASE 5. During what phase of the cell cycle does cell division occur? MITOSIS 6. During what phase of the cell cycle is DNA replicated? S-PHASE 7. During what phase of the cell cycle does the cell grow? G1,G2 8. During what phase of the cell cycle does the cell prepare for mitosis? G2 9. How many stages are there in mitosis? 4 10. Put the following stages of mitosis in order: anaphase, prophase, metaphase, and telophase. -

Cell Life Cycle and Reproduction the Cell Cycle (Cell-Division Cycle), Is a Series of Events That Take Place in a Cell Leading to Its Division and Duplication
Cell Life Cycle and Reproduction The cell cycle (cell-division cycle), is a series of events that take place in a cell leading to its division and duplication. The main phases of the cell cycle are interphase, nuclear division, and cytokinesis. Cell division produces two daughter cells. In cells without a nucleus (prokaryotic), the cell cycle occurs via binary fission. Interphase Gap1(G1)- Cells increase in size. The G1checkpointcontrol mechanism ensures that everything is ready for DNA synthesis. Synthesis(S)- DNA replication occurs during this phase. DNA Replication The process in which DNA makes a duplicate copy of itself. Semiconservative Replication The process in which the DNA molecule uncoils and separates into two strands. Each original strand becomes a template on which a new strand is constructed, resulting in two DNA molecules identical to the original DNA molecule. Gap 2(G2)- The cell continues to grow. The G2checkpointcontrol mechanism ensures that everything is ready to enter the M (mitosis) phase and divide. Mitotic(M) refers to the division of the nucleus. Cell growth stops at this stage and cellular energy is focused on the orderly division into daughter cells. A checkpoint in the middle of mitosis (Metaphase Checkpoint) ensures that the cell is ready to complete cell division. The final event is cytokinesis, in which the cytoplasm divides and the single parent cell splits into two daughter cells. Reproduction Cellular reproduction is a process by which cells duplicate their contents and then divide to yield multiple cells with similar, if not duplicate, contents. Mitosis Mitosis- nuclear division resulting in the production of two somatic cells having the same genetic complement (genetically identical) as the original cell. -
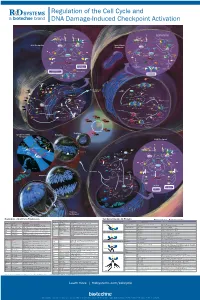
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

Meiosis I and Meiosis II; Life Cycles
Meiosis I and Meiosis II; Life Cycles Meiosis functions to reduce the number of chromosomes to one half. Each daughter cell that is produced will have one half as many chromosomes as the parent cell. Meiosis is part of the sexual process because gametes (sperm, eggs) have one half the chromosomes as diploid (2N) individuals. Phases of Meiosis There are two divisions in meiosis; the first division is meiosis I: the number of cells is doubled but the number of chromosomes is not. This results in 1/2 as many chromosomes per cell. The second division is meiosis II: this division is like mitosis; the number of chromosomes does not get reduced. The phases have the same names as those of mitosis. Meiosis I: prophase I (2N), metaphase I (2N), anaphase I (N+N), and telophase I (N+N) Meiosis II: prophase II (N+N), metaphase II (N+N), anaphase II (N+N+N+N), and telophase II (N+N+N+N) (Works Cited See) *3 Meiosis I (Works Cited See) *1 1. Prophase I Events that occur during prophase of mitosis also occur during prophase I of meiosis. The chromosomes coil up, the nuclear membrane begins to disintegrate, and the centrosomes begin moving apart. The two chromosomes may exchange fragments by a process called crossing over. When the chromosomes partially separate in late prophase, until they separate during anaphase resulting in chromosomes that are mixtures of the original two chromosomes. 2. Metaphase I Bivalents (tetrads) become aligned in the center of the cell and are attached to spindle fibers. -

Mitosis, Cytokinesis, Meiosis and Apoptosis - Michelle Gehringer
FUNDAMENTALS OF BIOCHEMISTRY, CELL BIOLOGY AND BIOPHYSICS – Vol. II - Mitosis, Cytokinesis, Meiosis and Apoptosis - Michelle Gehringer MITOSIS, CYTOKINESIS, MEIOSIS AND APOPTOSIS Michelle Gehringer Department of Biochemistry and Microbiology, University of Port Elizabeth, South Africa Keywords: Cell cycle, checkpoints, growth factors, mitosis, meiosis, cyclin, cyclin dependent protein kinases, G1 phase, S phase, spindle, prophase, anaphase, metaphase, telophase, cytokinesis, p53, apoptosis Contents 1. The eukaryote cell cycle 1.1. Phases 2. Mitosis 2.1 Prophase 2.2 Metaphase 2.3 Anaphase 2.4 Telophase 2.5 Cytokinesis 3. Meiosis 3.1. Stages of meiosis 4. Fertilization and development 5. Regulators of Cell cycle 5.1. Checkpoints 5.1.1 G1/S checkpoint 5.1.2 G2/M checkpoint 5.1.3 Mitosis checkpoint 5.2 Maturation promoting factor 5.3 Cyclin dependent protein kinases 5.3.1 Diversity and action 5.3.2 Regulation 5.3.3 Cyclin regulation of mitosis 5.4 Growth factors 5.5 Inhibitors of cell cycle progression 6. Programmed cell death 6.1. TriggersUNESCO of apoptosis – EOLSS 6.2. Pathways leading to apoptosis 7. Conclusion SAMPLE CHAPTERS Glossary Bibliography Biographical Sketch Summary The eukaryotic cell cycle comprises clear stages. Two major stages are the synthesis phase, where the cell replicates its genetic information, and the mitotic phase, where the cell divides into two daughter cells. They are separated by gap phases 1 and 2. These ©Encyclopedia of Life Support Systems (EOLSS) FUNDAMENTALS OF BIOCHEMISTRY, CELL BIOLOGY AND BIOPHYSICS – Vol. II - Mitosis, Cytokinesis, Meiosis and Apoptosis - Michelle Gehringer stages prepare the cell for the following step in the cell cycle. -

Mitotic Catastrophe
Cell Death and Differentiation (2008) 15, 1153–1162 & 2008 Nature Publishing Group All rights reserved 1350-9047/08 $30.00 www.nature.com/cdd Review Death through a tragedy: mitotic catastrophe H Vakifahmetoglu1, M Olsson1 and B Zhivotovsky*,1 Mitotic catastrophe (MC) has long been considered as a mode of cell death that results from premature or inappropriate entry of cells into mitosis and can be caused by chemical or physical stresses. Whereas it initially was depicted as the main form of cell death induced by ionizing radiation, it is today known to be triggered also by treatment with agents influencing the stability of microtubule, various anticancer drugs and mitotic failure caused by defective cell cycle checkpoints. Although various descriptions explaining MC exist, there is still no general accepted definition of this phenomenon. Here, we present evidences indicating that death-associated MC is not a separate mode of cell death, rather a process (‘prestage’) preceding cell death, which can occur through necrosis or apoptosis. The final outcome of MC depends on the molecular profile of the cell. Cell Death and Differentiation (2008) 15, 1153–1162; doi:10.1038/cdd.2008.47; published online 11 April 2008 Historical Remarks defective cell cycle checkpoints.8 MC was also depicted as the main form of cell death induced by ionizing radiation. During mitosis, proliferating cells undergo several structural The first observations of MC were made in the late 1930s and molecular changes, characterized by chromatin conden- and early 1940s when cells in exponential growth phase were sation, spindle formation, nuclear envelope fragmentation and exposed to radiation.9,10 It was noticed that the fraction of cells cytoskeleton reorganization.1 Chromosome segregation is in the mitotic stage instantly declined in response to radiation carried out by a complex machinery – the mitotic spindle – that and did not reappear until several hours following treatment. -

Ultrastructural Analysis of Intracellular Membrane and Microtubule Behavior During Mitosis of Drosophila S2 Cells
bioRxiv preprint doi: https://doi.org/10.1101/232199; this version posted December 11, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Ultrastructural analysis of intracellular membrane and microtubule behavior during mitosis of Drosophila S2 cells Anton Strunov1, 2, Lidiya V. Boldyreva1, Evgeniya N. Andreyeva1, Gera A. Pavlova1, Julia V. Popova1, 2, Alena V. Razuvaeva1, 3, Alina F. Anders1, 3, Fioranna Renda4, 5, Alexey V. Pindyurin1, 2, 3, Maurizio Gatti1, 4, Elena Kiseleva1, 2 1 Institute of Molecular and Cellular Biology, Siberian Branch of RAS, Novosibirsk, 630090, Russia 2 Institute of Cytology and Genetics, Siberian Branch of RAS, Novosibirsk, 630090, Russia 3 Novosibirsk State University, Novosibirsk, 630090, Russia 4 IBPM CNR and Department of Biology and Biotechnology, Sapienza University of Rome, Rome, 00185, Italy 5 Present address: Wadsworth Center, New York State Department of Health, Albany, NY 12201, USA Running Title: Ultrastructural analysis of S2 cell mitosis Keywords: S2 cells, Drosophila, nuclear membranes, spindle microtubules, kinetochores. Correspondence: Anton Strunov: [email protected] Maurizio Gatti: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/232199; this version posted December 11, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract S2 cells are one of the most widely used Drosophila melanogaster cell lines for molecular dissection of mitosis using RNA interference (RNAi). However, a detailed and complete description of S2 cell mitosis at the ultrastructural level is still missing.