Release of Stem Cells from Quiescence Reveals Multiple Gliogenic Domains in the Adult Brain
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Differential Expression of PDGFD in Cancers of the Breast
Differential expression of platelet-derived growth factor D in cancers of the breast. Shahan Mamoor, MS1 [email protected] East Islip, NY 11730 Breast cancer affects women at relatively high frequency1. We mined published microarray datasets2,3 to determine in an unbiased fashion and at the systems level genes most differentially expressed in the primary tumors of patients with breast cancer. We report here significant differential expression of the gene encoding platelet-derived growth factor D, PDGFD, when comparing primary tumors of the breast to the tissue of origin, the normal breast. PDGFD was also differentially expressed in the tumor cells of patients with triple negative breast cancer. PDGFD mRNA was present at significantly lower quantities in tumors of the breast as compared to normal breast tissue. Analysis of human survival data revealed that expression of PDGFD in primary tumors of the breast was correlated with overall survival in patients with basal and luminal A subtype cancer, demonstrating a relationship between primary tumor expression of a differentially expressed gene and patient survival outcomes influenced by molecular subtype. PDGFD may be of relevance to initiation, maintenance or progression of cancers of the female breast. Keywords: breast cancer, PDGFD, platelet-derived growth factor D, systems biology of breast cancer, targeted therapeutics in breast cancer. 1 Invasive breast cancer is diagnosed in over a quarter of a million women in the United States each year1 and in 2018, breast cancer was the leading cause of cancer death in women worldwide4. While patients with localized breast cancer are provided a 99% 5-year survival rate, patients with regional breast cancer, cancer that has spread to lymph nodes or nearby structures, are provided an 86% 5-year survival rate5,6. -

PDGF-C and PDGF-D Signaling in Vascular Diseases and Animal Models
Molecular Aspects of Medicine 62 (2018) 1e11 Contents lists available at ScienceDirect Molecular Aspects of Medicine journal homepage: www.elsevier.com/locate/mam PDGF-C and PDGF-D signaling in vascular diseases and animal models * Erika Folestad a, Anne Kunath b, Dick Wågsater€ b, a Division of Vascular Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden b Division of Drug Research, Department of Medical and Health Sciences, Linkoping€ University, Linkoping,€ Sweden article info abstract Article history: Members of the platelet-derived growth factor (PDGF) family are well known to be involved in different Received 31 August 2017 pathological conditions. The cellular and molecular mechanisms induced by the PDGF signaling have Received in revised form been well studied. Nevertheless, there is much more to discover about their functions and some 14 November 2017 important questions to be answered. This review summarizes the known roles of two of the PDGFs, Accepted 22 January 2018 PDGF-C and PDGF-D, in vascular diseases. There are clear implications for these growth factors in several Available online 14 February 2018 vascular diseases, such as atherosclerosis and stroke. The PDGF receptors are broadly expressed in the cardiovascular system in cells such as fibroblasts, smooth muscle cells and pericytes. Altered expression Keywords: Aneurysm of the receptors and the ligands have been found in various cardiovascular diseases and current studies fi Atherosclerosis have shown important implications of PDGF-C and PDGF-D signaling in brosis, neovascularization, Growth factor atherosclerosis and restenosis. Myocardial infarction © 2018 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND Smooth muscle cells license (http://creativecommons.org/licenses/by-nc-nd/4.0/). -

Xo PANEL DNA GENE LIST
xO PANEL DNA GENE LIST ~1700 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes (at 400 -500x average coverage on tumor) Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 -

Differences in Gene Expression Profiles and Carcinogenesis Pathways Involved in Cisplatin Resistance of Four Types of Cancer
596 ONCOLOGY REPORTS 30: 596-614, 2013 Differences in gene expression profiles and carcinogenesis pathways involved in cisplatin resistance of four types of cancer YONG YANG1,2, HUI LI1,2, SHENGCAI HOU1,2, BIN HU1,2, JIE LIU1,3 and JUN WANG1,3 1Beijing Key Laboratory of Respiratory and Pulmonary Circulation, Capital Medical University, Beijing 100069; 2Department of Thoracic Surgery, Beijing Chao-Yang Hospital, Capital Medical University, Beijing 100020; 3Department of Physiology, Capital Medical University, Beijing 100069, P.R. China Received December 23, 2012; Accepted March 4, 2013 DOI: 10.3892/or.2013.2514 Abstract. Cisplatin-based chemotherapy is the standard Introduction therapy used for the treatment of several types of cancer. However, its efficacy is largely limited by the acquired drug Cisplatin is primarily effective through DNA damage and is resistance. To date, little is known about the RNA expression widely used for the treatment of several types of cancer, such changes in cisplatin-resistant cancers. Identification of the as testicular, lung and ovarian cancer. However, the ability RNAs related to cisplatin resistance may provide specific of cancer cells to become resistant to cisplatin remains a insight into cancer therapy. In the present study, expression significant impediment to successful chemotherapy. Although profiling of 7 cancer cell lines was performed using oligo- previous studies have identified numerous mechanisms in nucleotide microarray analysis data obtained from the GEO cisplatin resistance, it remains a major problem that severely database. Bioinformatic analyses such as the Gene Ontology limits the usefulness of this chemotherapeutic agent. Therefore, (GO) and KEGG pathway were used to identify genes and it is crucial to examine more elaborate mechanisms of cisplatin pathways specifically associated with cisplatin resistance. -

Factors Secreted by Cancer-Associated Fibroblasts That Sustain Cancer Stem Properties in Head and Neck Squamous Carcinoma Cells As Potential Therapeutic Targets
cancers Article Factors Secreted by Cancer-Associated Fibroblasts that Sustain Cancer Stem Properties in Head and Neck Squamous Carcinoma Cells as Potential Therapeutic Targets Saúl Álvarez-Teijeiro 1,2,†,* , Cristina García-Inclán 1,†, M. Ángeles Villaronga 1,2, Pedro Casado 3 , Francisco Hermida-Prado 1 , Rocío Granda-Díaz 1, Juan P. Rodrigo 1,2 , Fernando Calvo 4, Nagore del-Río-Ibisate 1, Alberto Gandarillas 5, Francisco Morís 6, Mario Hermsen 1,2, Pedro Cutillas 3 and Juana M. García-Pedrero 1,2,* 1 Department of Otolaryngology, Hospital Universitario Central de Asturias and Instituto de Investigación Sanitaria del Principado de Asturias; Instituto Universitario de Oncología del Principado de Asturias, University of Oviedo, 33011 Oviedo, Spain; [email protected] (C.G.-I.); [email protected] (M.Á.V.); [email protected] (F.H.-P.); [email protected] (R.G.-D.); [email protected] (J.P.R.); [email protected] (N.d.-R.-I.); [email protected] (M.H.) 2 CIBERONC, 28029 Madrid, Spain 3 Cell Signalling & Proteomics Group, Barts Cancer Institute, Queen Mary University of London, London EC1M 6BQ, UK; [email protected] (P.Ca.); [email protected] (P.Cu.) 4 Tumour Microenvironment Team, Division of Cancer Biology, Institute of Cancer Research, 237 Fulham Road, London SW3 6JB, UK; [email protected] 5 Cell Cycle, Stem Cell Fate and Cancer Lab Instituto de Investigación Marqués de Valdecilla (IDIVAL), 39011 Santander, Spain; [email protected] 6 EntreChem SL, Vivero Ciencias de la Salud, 33011 Oviedo, Spain; [email protected] * Correspondence: [email protected] (S.Á.-T.); juanagp.fi[email protected] (J.M.G.-P.) † These authors contributed equally to this work. -
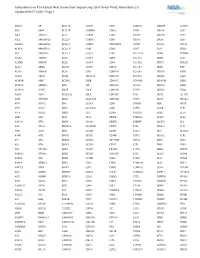
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1
Comprehensive Pan-Cancer Next Generation Sequencing Solid Tumor Panel, Aberration List Updated 08-07-2020 --Page 1 ABCC3 AR BCL11A CANT1 CDK1 CMKLR1 DAB2IP DUSP9 ABI1 ARAF BCL11B CAPRIN1 CDK12 CNBP DACH1 E2F1 ABL1 ARFRP1 BCL2 CAPZB CDK2 CNOT2 DACH2 E2F3 ABL2 ARHGAP20 BCL2A1 CARD11 CDK4 CNTN1 DAXX EBF1 ABLIM1 ARHGAP26 BCL2L1 CARM1 CDK5RAP2 CNTRL DCLK2 ECT2L ACACA ARHGEF12 BCL2L11 CARS CDK6 COG5 DCN EDIL3 ACE ARHGEF7 BCL2L2 CASC5 CDK7 COL11A1 DDB1 EDNRB ACER1 ARID1A BCL3 CASP3 CDK8 COL1A1 DDB2 EED ACSBG1 ARID1B BCL6 CASP7 CDK9 COL1A2 DDIT3 EEFSEC ACSL3 ARID2 BCL7A CASP8 CDKL5 COL3A1 DDR2 EGF ACSL6 ARID5B BCL9 CAV1 CDKN1A COL6A3 DDX10 EGFR ACVR1 ARIH2 BCOR CBFA2T3 CDKN1B COL9A3 DDX20 EGR1 ACVR1B ARNT BCORL1 CBFB CDKN1C COMMD1 DDX39B EGR2 ACVR1C ARRDC4 BCR CBL CDKN2A COX6C DDX3X EGR3 ACVR2A ASMTL BDNF CBLB CDKN2B CPNE1 DDX41 EGR4 ADD3 ASPH BHLHE22 CBLC CDKN2C CPS1 DDX5 EIF1AX ADM ASPSCR1 BICC1 CCDC28A CDKN2D CPSF6 DDX6 EIF4A2 AFF1 ASTN2 BIN1 CCDC6 CDX1 CRADD DEK EIF4E AFF3 ASXL1 BIRC3 CCDC88C CDX2 CREB1 DGKB ELF3 AFF4 ASXL2 BIRC6 CCK CEBPA CREB3L1 DGKI ELF4 AGR3 ATF1 BLM CCL2 CEBPB CREB3L2 DGKZ ELK4 AHCYL1 ATF3 BMP4 CCNA2 CEBPD CREBBP DICER1 ELL AHI1 ATG13 BMPR1A CCNB1IP1 CEBPE CRKL DIRAS3 ELN AHR ATG5 BRAF CCNB3 CENPF CRLF2 DIS3 ELOVL2 AHRR ATIC BRCA1 CCND1 CENPU CRTC1 DIS3L2 ELP2 AIP ATL1 BRCA2 CCND2 CEP170B CRTC3 DKK1 EML1 AK2 ATM BRCC3 CCND3 CEP57 CSF1 DKK2 EML4 AK5 ATP1B4 BRD1 CCNE1 CEP85L CSF1R DKK4 ENPP2 AKAP12 ATP8A2 BRD3 CCNG1 CHCHD7 CSF3 DLEC1 EP300 AKAP6 ATR BRD4 CCT6B CHD2 CSF3R DLL1 EP400 AKAP9 ATRNL1 BRIP1 CD19 CHD4 CSNK1A1 DLL3 -

The Hif1α-PDGFD-Pdgfrα Axis Controls Glioblastoma Growth At
Peng et al. Journal of Experimental & Clinical Cancer Research (2021) 40:278 https://doi.org/10.1186/s13046-021-02082-7 RESEARCH Open Access The HIF1α-PDGFD-PDGFRα axis controls glioblastoma growth at normoxia/mild- hypoxia and confers sensitivity to targeted therapy by echinomycin Gong Peng1†, Yin Wang2†, Pengfei Ge3, Christopher Bailey2, Peng Zhang4, Di Zhang5, Zhaoli Meng1, Chong Qi1, Qian Chen1, Jingtao Chen1, Junqi Niu1, Pan Zheng2,6, Yang Liu2,6* and Yan Liu2* Abstract Background: Glioblastoma multiforme (GBM), a lethal brain tumor, remains the most daunting challenge in cancer therapy. Overexpression and constitutive activation of PDGFs and PDGFRα are observed in most GBM; however, available inhibitors targeting isolated signaling pathways are minimally effective. Therefore, better understanding of crucial mechanisms underlying GBM is needed for developing more effective targeted therapies. Methods: Target genes controlled by HIF1α in GBM were identified by analysis of TCGA database and by RNA- sequencing of GBM cells with HIF1α knockout by sgRNA-Cas9 method. Functional roles of HIF1α, PDGFs and PDGF Rs were elucidated by loss- or gain-of-function assays or chemical inhibitors, and compared in response to oxygen tension. Pharmacological efficacy and gene expression in mice with intracranial xenografts of primary GBM were analyzed by bioluminescence imaging and immunofluorescence. Results: HIF1α binds the PDGFD proximal promoter and PDGFRA intron enhancers in GBM cells under normoxia or mild-hypoxia to induce their expression and maintain constitutive activation of AKT signaling, which in turn increases HIF1α protein level and activity. Paradoxically, severe hypoxia abrogates PDGFRα expression despite enhancing HIF1α accumulation and corresponding PDGF-D expression. -

SUPPLEMENTARY APPENDIX Exome Sequencing Reveals Heterogeneous Clonal Dynamics in Donor Cell Myeloid Neoplasms After Stem Cell Transplantation
SUPPLEMENTARY APPENDIX Exome sequencing reveals heterogeneous clonal dynamics in donor cell myeloid neoplasms after stem cell transplantation Julia Suárez-González, 1,2 Juan Carlos Triviño, 3 Guiomar Bautista, 4 José Antonio García-Marco, 4 Ángela Figuera, 5 Antonio Balas, 6 José Luis Vicario, 6 Francisco José Ortuño, 7 Raúl Teruel, 7 José María Álamo, 8 Diego Carbonell, 2,9 Cristina Andrés-Zayas, 1,2 Nieves Dorado, 2,9 Gabriela Rodríguez-Macías, 9 Mi Kwon, 2,9 José Luis Díez-Martín, 2,9,10 Carolina Martínez-Laperche 2,9* and Ismael Buño 1,2,9,11* on behalf of the Spanish Group for Hematopoietic Transplantation (GETH) 1Genomics Unit, Gregorio Marañón General University Hospital, Gregorio Marañón Health Research Institute (IiSGM), Madrid; 2Gregorio Marañón Health Research Institute (IiSGM), Madrid; 3Sistemas Genómicos, Valencia; 4Department of Hematology, Puerta de Hierro General University Hospital, Madrid; 5Department of Hematology, La Princesa University Hospital, Madrid; 6Department of Histocompatibility, Madrid Blood Centre, Madrid; 7Department of Hematology and Medical Oncology Unit, IMIB-Arrixaca, Morales Meseguer General University Hospital, Murcia; 8Centro Inmunológico de Alicante - CIALAB, Alicante; 9Department of Hematology, Gregorio Marañón General University Hospital, Madrid; 10 Department of Medicine, School of Medicine, Com - plutense University of Madrid, Madrid and 11 Department of Cell Biology, School of Medicine, Complutense University of Madrid, Madrid, Spain *CM-L and IB contributed equally as co-senior authors. Correspondence: -

PDGFR) Inhibitors in the Treatment of Neoplastic Disorders
Pharmacological Research 129 (2018) 65–83 Contents lists available at ScienceDirect Pharmacological Research j ournal homepage: www.elsevier.com/locate/yphrs Invited Review The role of small molecule platelet-derived growth factor receptor (PDGFR) inhibitors in the treatment of neoplastic disorders Robert Roskoski Jr. Blue Ridge Institute for Medical Research, 3754 Brevard Road, Suite 116, Box 19, Horse Shoe, NC, 28742-8814, United States a r t i c l e i n f o a b s t r a c t Article history: Platelet-derived growth factor (PDGF) was discovered as a serum-derived component necessary for the Received 28 January 2018 growth of smooth muscle cells, fibroblasts, and glial cells. The PDGF family is a product of four gene prod- Accepted 29 January 2018 ucts and consists of five dimeric isoforms: PDGF-AA, PDGF-BB, PDGF-CC, PDGF-DD, and the PDGF-AB Available online 3 February 2018 heterodimer. This growth factor family plays an essential role in embryonic development and in wound healing in the adult. These growth factors mediate their effects by binding to and activating their receptor Chemical compounds studied in this article: protein-tyrosine kinases, which are encoded by two genes: PDGFRA and PDGFRB. The functional recep- Axitinib: (PubMED CID: 6450551) tors consist of the PDGFR␣/␣ and PDGFR/ homodimers and the PDGFR␣/ heterodimer. Although Dasatinib: (PubMED CID: 3062316) PDGF signaling is most closely associated with mesenchymal cells, PDGFs and PDGF receptors are widely Imatinib: (PubMED CID: 5291) expressed in the mammalian central nervous system. The PDGF receptors contain an extracellular domain Lenvatinib: (PubMED CID: 9823820) d Nilotinib: (PubMed CID: 644241) that is made up of five immunoglobulin-like domains (Ig- 1/2/3/4/5), a transmembrane segment, a jux- Nintedanib: (PubMed CID: 9809715) tamembrane segment, a protein-tyrosine kinase domain that contains an insert of about 100 amino acid Ponatinib: (PubMed CID: 24826799) residues, and a carboxyterminal tail. -

Tumor-Driven Paracrine Platelet-Derived Growth Factor Receptor a Signaling Is a Key Determinant of Stromal Cell Recruitment Inamodelofhumanlungcarcinoma Max L
Human Cancer Biology Tumor-Driven Paracrine Platelet-Derived Growth Factor Receptor A Signaling Is a Key Determinant of Stromal Cell Recruitment inaModelofHumanLungCarcinoma Max L. Tejada,1Lanlan Yu,1Jianying Dong,1Kenneth Jung,3 Gloria Meng,4 Franklin V. Peale,2 Gretchen D. Frantz,2 Linda Hall,2 XiaoHuan Liang,1Hans-Peter Gerber,1and Napoleone Ferrara1 Abstract Activated fibroblasts are thought to play important roles in the progression of many solid tumors, but little is known about the mechanisms responsible for the recruitment of fibroblasts in tumors. Using several methods, we identified platelet-derived growth factor A (PDGFA) as the major fibroblast chemoattractant and mitogen from conditioned medium generated by the Calu-6 lung carcinoma cell line. In addition, we showed that Calu-6 tumors express significant levels of PDGFC, and that the levels of expression of these two PDGFRa ligands correlate strongly with the degree of stromal fibroblast infiltration into the tumor mass. The most intense expression of PDGFRa was observed in fibroblasts in the tumor outer rim. We subsequently showed that disrupting PDGFRa-mediated signaling results in significant inhibition of tumor growth in vivo. Furthermore, analysis of a compendium of microarray data revealed significant expression of PDGFA, PDGFC, and PDGFRa in human lung tumors. We propose that therapies targeting this stromal cell type may be effective in treating certain types of solid tumors. It has become apparent that the microenvironment in which markersconsistentwiththosefoundatthesitesofnormal tumor cells develop profoundly influences many steps of tumor wound healing (3). progression. The tumor microenvironment consists of a stroma, In the normal wound repair process, fibroblasts are recruited which is composed of immune and inflammatory cells, from the surrounding tissue. -

Original Article Effect of Mesenchymal Stem Cell Derived Exosomes Carrying PDGFD on Lung Cancer
Int J Clin Exp Pathol 2017;10(1):224-232 www.ijcep.com /ISSN:1936-2625/IJCEP0041431 Original Article Effect of mesenchymal stem cell derived exosomes carrying PDGFD on lung cancer Feng Huang1, Yongliang Yao1, Jianhong Wu1, Liya Yu1, Shiyue Wu1, Xiongyong Pu1, Liang Xu1, Mei Wang2, Longfei Xia2 1Department of Clinical Laboratory, The First People’s Hospital of Kunshan Affiliated with Jiangsu University, Su- zhou, China; 2School of Medicine, Jiangsu University, Zhenjiang, China Received October 9, 2016; Accepted October 25, 2016; Epub January 1, 2017; Published January 15, 2017 Abstract: Mesenchymal stem cells (MSCs) are capable of promoting lung tumor progression, but the underlying mechanism remains unclear. In this study, we investigated the effect of bone marrow MSC-derived exosomes (MSC- exosomes) carrying PDGFD on in vitro and in vivo lung tumor growth and its underlying mechanism. MSC-exosomes carrying PDGFD promoted lung cancer cell migration, EMT, proliferation and PI3K signaling pathway. Recombinant PDGFD could mimic the effect of MSC-exosomes carrying PDGFD on lung cancer cells. These effects were abol- ished by a neutralizing antibody against PDGFD in vitro and in vivo. We found that MSC-exosomes were capable of regulating lung cancer cell progression through PDGFD MSC-exosomes carry. This study may pave the way for novel therapeutic strategy targeting the PDGFD pathway regulating MSC-exosome-mediated cell-cell interactions. Keywords: MSC, exosome, PDGFD, lung cancer cells Introduction and MSCs [10, 11]. Studies have shown that paracrine signals from MSCs significantly in- Non-small cell lung cancer (NSCLC) is the lead- fluence tumor cell behavior [12, 13]; however, ing cause of cancer-related mortality world- the mechanism underlying tumor progression wide. -

Genequery™ Human Growth Factors Qpcr Array Kit (GQH-GRF) Catalog #GK123 Product
GeneQuery™ Human Growth Factors qPCR Array Kit (GQH-GRF) Catalog #GK123 Product Description ScienCell's GeneQuery™ Human Growth Factors qPCR Array Kit (GQH-GRF) is designed to facilitate gene expression profiling of 88 human growth factor genes. Growth factors are proteins or hormones which are critical in regulating cellular processes such as apoptosis, proliferation, differentiation, inflammation and wound healing. Brief examples of how genes may be grouped according to their functions are shown below: • Angiopoietins: ANGPT1, ANGPT2 • Bone morphogenetic proteins: BMP1, BMP10, BMP2, BMP4, BMP6, BMP7, BMP8B • Ciliary neurotrophic factor family: CNTF, LIF, IL6 • Colony-stimulating factors: CSF1, CSF2, CSF3 • Epidermal growth factor: EGF • Ephrins: EFNA1, EFNA2, EFNA3, EFNA4, EFNA5, EFNB1, EFNB2, EFNB3 • Fibroblast growth factors: FGF1, FGF10, FGF13, FGF14, FGF16, FGF17, FGF18, FGF19, FGF2, FGF20, FGF23, FGF3, FGF4, FGF5, FGF7, FGF8, FGF9 • GDNF family: GDNF, NRTN, ARTN, PSPN • Growth differentiation factors: GDF1, GDF10, GDF11, GDF2, GDF3, MSTN • Hepatocyte growth factor: HGF • Insulin-like growth factors: IGF1, IGF2 • Interleukins: IL1A, IL1B, IL2, IL3, IL4, IL5, IL6, IL7, IL9, IL10, IL11, IL12B, IL15, IL18 • Neuregulins: NRG1, NRG3 • Neurotrophins: BDNF, NGF, NTF3, NTF4 • Platelet-derived growth factors: PDGFA, PDGFB, PDGFC, PDGFD • Thrombopoietin: THPO • Transforming growth factors: TGFA, TGFB1, TGFB2, TGFB3 • Tumor necrosis factor: TNF • Vascular endothelial growth factors: VEGFA, VEGFB, VEGFC, VEGFD, PGF Note : all gene names follow their official symbols by the Human Genome Organization Gene Nomenclature Committee (HGNC). GeneQuery™ qPCR array kits are qPCR ready in a 96-well plate format, with each well containing one primer set that can specifically recognize and efficiently amplify a target gene's cDNA.