Dense Tracking of the Dynamics of the Microbial Community and Chemicals Constituents in Spontaneous Wheat Sourdough During Two Months of Backslopping
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Autumn Conference Proceedings 2015 British Society of Baking
2FWREHU $XWXPQ&RQIHUHQFH 3URFHHGLQJV %ULWLVK6RFLHW\RI%DNLQJ $IILOLDWHGWRWKH$PHULFDQ6RFLHW\RI%DNLQJ %ULWLVK6RFLHW\RI%DNLQJ $XWXPQ&RQIHUHQFH %LFHVWHU+RWHO*ROIDQG6SD2[IRUGVKLUH 7XHVGD\WKDQG:HGQHVGD\WK2FWREHU 3DSHU 6SHDNHU 3DJH 7KH%6%²3DVW3UHVHQWDQG)XWXUH -LP%URZQ 3DXO7XUQHU 3 0LNH%DJVKDZ 0D[LPLVLQJ%XVLQHVVDQG+XPDQ3RWHQWLDO 'DYLG6PDUW 8 :DVWH0DQDJHPHQWLQ)RRG0DQXIDFWXUH &DPSEHOO0XUUD\ 13 7KH&XUUHQW8.(FRQRPLF&OLPDWHIRU%XVLQHVV 'DQLHO/HH 19 7KH%,$5LVLQJ6WDU$ZDUG 1DWKDQ*LOHV 23 )UHHIURP'HYHORSPHQW %DNHU\3URGXFW,QQRYDWLRQ&KULV%URFNPDQ 27 7KH9LOODJH%DNHU\·V7UDLQLQJDQG,QQRYDWLRQ$FDGHP\5RELQ-RQHV 32 7KH6HFUHWDU\ %ULWLVK6RFLHW\RI%DNLQJ 9LQH&RWWDJH7RPSNLQV/DQH0DUVK*LEERQ %LFHVWHU2[RQ2;(; 7HO)D[(PDLOEVE#IUHHXNFRP 1 7+(%5,7,6+62&,(7<2)%$.,1* $IILOLDWHGWRWKH$PHULFDQ6RFLHW\RI%DNLQJ ([HFXWLYH&RPPLWWHH 0LNH%DJVKDZ &KDLUPDQ %ULDQ&ODUNH 3DXO7XUQHU 9LFH&KDLUPDQ 5LFKDUG+D]HOGLQH -LP%URZQ +RQ7UHDVXUHU 6\OYLD0DFGRQDOG &RQIHUHQFH&RRUGLQDWRU *RUGRQ3ROVRQ 6KDURQ%\UQH 6HFUHWDU\ -DQH7\OHU 6DUD$XWWRQ ,PPHGLDWH3DVW&KDLUPDQ 3DXO:HVWRQ 6DUD3ULHVWOH\ 6WXGHQW/LDLVRQ 0DLO&KLPS 0DUN<RXQJ 3DVW&KDLUPHQ -7KRPVRQ $%XFKDQDQ 5)HUJXVRQ +&ROERXUQH )(OOLV 3+H\JDWH )%DWHV *3ULQFH 6&DXYDLQ $+DOO 32UW 10HDGRZV -6WHYHQV '5REHUWV &/RPD[ 36KHQWRQ 51HZVWHDG 1-DFNVRQ *+XPSKUH\ 70RVV $03ROODUG :*XQVWRQH 'U3:RRG .+RXOLVWRQ '.LQJ 30DVVH\ 6$XWWRQ 5+DUH $:DWHUILHOG 7%HDOH $+RGJHV 5+XPSKUH\ -%URZQ -5LWFKLH 3:DUG 5+RUQVE\ -53DUNLQVRQ -$QWKRQ\ ,0HOOLQJ *9HUH .6KDZ '*DUUDWW 56LPPV )6D\HU *&ULWLFRV )(OOLV 30RUURZ 5)OLQW 5.LUN .6\GQH\ -*ULHYHV 3DVWQG9LFH&KDLUPHQ EHIRUHWKHQG9LFH&KDLUPDQGLGQRWSURFHHGWR VW9LFH&KDLUPDQDQG&KDLUPDQEHFDXVHKHGLGQRWZRUNIRUDEDNHU\FRPSDQ\ 36DYRU\ 57XUQHU *%UXFH .0RUJDQ -0DKOLFK .&ROOLQJH **LOEHUW 1'RXJODV 7&ROOLQV -3ULFH 6/DPEHUW -%URZQ 1%HVVDQW -+X[WDEOH ::DOODFH 56DQGHUVRQ :3ULQJOH *6FKLQGOHU -3HONPDQ '(OLDV &&XUWLV ,.LQJ .:LOOLDPVRQ -3ULQJOH &%UDFHZHOO -*UHHQILHOG 2 Paul Turner; and to then look to the future with Chairman Mike 2015 DIAMOND JUBILEE AUTUMN Bagshaw. -

Tokyo Food Technology Week 2021
Exhibit Brochure Tokyo Food Technology Week 2021 Sep. 2-3 2021 Sep. 2-3 2021 10:00~17:00 PACIFICO YOKOHAMA Exhibition Hall C [Website] https://tokyofoodtechnology.com/2021/en.pdf Organizer:EJK Japan, Ltd./ The Japan Food News Show profile The Gateway into Japan Market ! Tokyo Food Technology Week “Tokyo Food Technology Week” will be expandingly held by including “P & B JAPAN”, “Nutritious Food Ingredients EXPO”, and “food ingredients EXPO for Pre-packaged, Frozen, and Instant food”. TFTW creates the synergistic effect of 3 shows by proposing the cutting- edge technology that shows new trends in the whole food industry. LOGO nd NAME OF 2 Nutritious Food 1st food ingredients EXPO for Pre- 9th P&B JAPAN (P&B) SHOW Ingredients EXPO (NFI) packaged, Frozen, and Instant food (PFI) ORGANIZER(S) EJK Japan, Ltd. EJK Japan, Ltd./ The Japan Food News Food ingredients for bread Health functional food Food Ingredients (meat, & confectionery (flour, oil, ingredients (vitamin & sea food, vegetable, fruit, yeast, jam, honey, frozen mineral, oligo sugar, food cereal, seasoning, spice, dough, etc. ) / Food fiber, lactic acid bacteria, herb etc.) / Food additives ingredients for sandwich, sugar alcohol, protein, / Professional products for EXHBITOR burger (meat, vegetable, amino acid, enzyme, etc.) restaurant chain / Food PROFILE dairy product, sauce, etc. ) Vegetable / Herb / Honey processing technology / Coffee & tea & beverages product / Soy product / (freezing, thawing, / Kitchen equipment / POS Component analysis / sterilizer drying etc. )/ register -

Characterization of Weissella Koreensis SK Isolated From
microorganisms Article Characterization of Weissella koreensis SK Isolated from Kimchi Fermented at Low Temperature ◦ (around 0 C) Based on Complete Genome Sequence and Corresponding Phenotype So Yeong Mun and Hae Choon Chang * Department of Food and Nutrition, Kimchi Research Center, Chosun University, 309 Pilmun-daero, Dong-gu, Gwangju 61452, Korea; [email protected] * Correspondence: [email protected] Received: 17 June 2020; Accepted: 28 July 2020; Published: 29 July 2020 Abstract: This study identified lactic acid bacteria (LAB) that play a major role in kimchi fermented at low temperature, and investigated the safety and functionality of the LAB via biologic and genomic analyses for its potential use as a starter culture or probiotic. Fifty LAB were isolated from 45 kimchi samples fermented at 1.5~0 C for 2~3 months. Weissella koreensis strains were determined as the − ◦ dominant LAB in all kimchi samples. One strain, W. koreensis SK, was selected and its phenotypic and genomic features characterized. The complete genome of W. koreensis SK contains one circular chromosome and plasmid. W. koreensis SK grew well under mesophilic and psychrophilic conditions. W. koreensis SK was found to ferment several carbohydrates and utilize an alternative carbon source, the amino acid arginine, to obtain energy. Supplementation with arginine improved cell growth and resulted in high production of ornithine. The arginine deiminase pathway of W. koreensis SK was encoded in a cluster of four genes (arcA-arcB-arcD-arcC). No virulence traits were identified in the genomic and phenotypic analyses. The results indicate that W. koreensis SK may be a promising starter culture for fermented vegetables or fruits at low temperature as well as a probiotic candidate. -

Annual Investment Report 2015-2016
South Carolina Retirement System Investment Commission 2015-2016 Annual Investment Report Photos courtesy of the Governor’s Office and the SC National Guard. South Carolina Retirement System Investment Commission Annual Investment Report Fiscal Year Ended June 30, 2016 Capitol Center 1201 Main Street, Suite 1510 Columbia, SC 29201 Edward Giobbe, MBA Chair For the period July 1, 2014 - June 30, 2016 Table of Contents Chair Report .....................................................................................................1 Consultant’s Letter ............................................................................................3 Overview ...........................................................................................................5 Commission ......................................................................................................6 Policy Allocation ...............................................................................................9 Manager Returns (Net of Fees) .......................................................................10 Securities Lending ..........................................................................................14 Expenses ..........................................................................................................15 Risk ..................................................................................................................17 Appendix Appendix A: Summary Schedule of Assets and Derivatives .....................19 Appendix B: Cash & -

Plant-Based Alternatives to Yogurt: State-Of-The-Art and Perspectives of New Biotechnological Challenges
foods Review Plant-Based Alternatives to Yogurt: State-of-the-Art and Perspectives of New Biotechnological Challenges Marco Montemurro 1 , Erica Pontonio 1 , Rossana Coda 2,3 and Carlo Giuseppe Rizzello 4,* 1 Department of Soil, Plant, and Food Science, University of Bari Aldo Moro, 70126 Bari, Italy; [email protected] (M.M.); [email protected] (E.P.) 2 Department of Food and Nutrition, University of Helsinki, 00014 Helsinki, Finland; rossana.coda@helsinki.fi 3 Helsinki Institute of Sustainability Science, 00014 Helsinki, Finland 4 Department of Environmental Biology, “Sapienza” University of Rome, 00185 Rome, Italy * Correspondence: [email protected] Abstract: Due to the increasing demand for milk alternatives, related to both health and ethical needs, plant-based yogurt-like products have been widely explored in recent years. With the main goal to obtain snacks similar to the conventional yogurt in terms of textural and sensory properties and ability to host viable lactic acid bacteria for a long-time storage, several plant-derived ingredients (e.g., cereals, pseudocereals, legumes, and fruits) as well as technological solutions (e.g., enzymatic and thermal treatments) have been investigated. The central role of fermentation in yogurt-like production led to specific selections of lactic acid bacteria strains to be used as starters to guarantee optimal textural (e.g., through the synthesis of exo-polysaccharydes), nutritional (high protein digestibility and low content of anti-nutritional compounds), and functional (synthesis of bioactive compounds) features of the products. This review provides an overview of the novel insights on fermented yogurt-like products. The state-of-the-art on the use of unconventional ingredients, traditional and innovative biotechnological processes, and the effects of fermentation on the textural, Citation: Montemurro, M.; nutritional, functional, and sensory features, and the shelf life are described. -

MOS 09 Financial Data
Consolidated Balance Sheets March 31, 2009 and 2008 (Thousands of yen) (Thousands of yen) 2009 2008 2009 2008 <Assets><Liabilities> Current Assets 17,616,897 16,683,196 Current Liabilities 8,456,486 8,029,799 Cash and deposits 6,760,422 7,878,058 Trade accounts payable 4,436,125 3,391,506 Accounts receivable 3,831,833 4,391,045 Income taxes payable 171,016 258,372 Marketable securities 2,147,624 1,049,879 Reserve for bonus payments 365,499 387,918 Merchandise, Supplies 3,256,827 2,197,766 Other 3,483,844 3,992,001 Deferred tax assets 115,946 - Fixed Liabilities 1,663,277 2,224,011 Other 1,670,044 1,605,866 Severance and retirement benefits for employees 70,816 37,660 Allowance for doubtful accounts -165,800 -439,419 Other 1,592,460 2,186,351 Total Liabilities 10,119,763 10,253,810 Fixed Assets 27,057,667 28,795,985 Property and Equipment 8,438,526 10,333,466 <Shareholders' Equity> Buildings 4,933,363 5,854,114 Common stock 11,412,845 11,412,845 Land 2,017,226 2,178,626 Capital surplus 11,100,524 11,100,524 Other 1,487,937 2,300,726 Retained earnings 14,185,797 13,832,667 Intangible Fixed Assets 1,187,322 1,535,893 Treasury Stock -1,604,182 -1,311,850 Investments and Other Assets 17,431,818 16,926,625 Net unrealised holding Gain(Loss)on Securities -426,094 17,858 Investments in securities 6,506,651 5,429,656 Foreign currency translation adjustments -332,975 -38,959 Deferred tax assets 603,748 788,737 Retained earning brought forward Other 10,698,174 11,120,910 Total net assets 34,554,802 35,225,371 Allowance for doubtful accounts -376,757 -
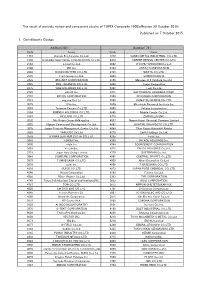
Published on 7 October 2015 1. Constituents Change the Result Of
The result of periodic review and component stocks of TOPIX Composite 1500(effective 30 October 2015) Published on 7 October 2015 1. Constituents Change Addition( 80 ) Deletion( 72 ) Code Issue Code Issue 1712 Daiseki Eco.Solution Co.,Ltd. 1972 SANKO METAL INDUSTRIAL CO.,LTD. 1930 HOKURIKU ELECTRICAL CONSTRUCTION CO.,LTD. 2410 CAREER DESIGN CENTER CO.,LTD. 2183 Linical Co.,Ltd. 2692 ITOCHU-SHOKUHIN Co.,Ltd. 2198 IKK Inc. 2733 ARATA CORPORATION 2266 ROKKO BUTTER CO.,LTD. 2735 WATTS CO.,LTD. 2372 I'rom Group Co.,Ltd. 3004 SHINYEI KAISHA 2428 WELLNET CORPORATION 3159 Maruzen CHI Holdings Co.,Ltd. 2445 SRG TAKAMIYA CO.,LTD. 3204 Toabo Corporation 2475 WDB HOLDINGS CO.,LTD. 3361 Toell Co.,Ltd. 2729 JALUX Inc. 3371 SOFTCREATE HOLDINGS CORP. 2767 FIELDS CORPORATION 3396 FELISSIMO CORPORATION 2931 euglena Co.,Ltd. 3580 KOMATSU SEIREN CO.,LTD. 3079 DVx Inc. 3636 Mitsubishi Research Institute,Inc. 3093 Treasure Factory Co.,LTD. 3639 Voltage Incorporation 3194 KIRINDO HOLDINGS CO.,LTD. 3669 Mobile Create Co.,Ltd. 3197 SKYLARK CO.,LTD 3770 ZAPPALLAS,INC. 3232 Mie Kotsu Group Holdings,Inc. 4007 Nippon Kasei Chemical Company Limited 3252 Nippon Commercial Development Co.,Ltd. 4097 KOATSU GAS KOGYO CO.,LTD. 3276 Japan Property Management Center Co.,Ltd. 4098 Titan Kogyo Kabushiki Kaisha 3385 YAKUODO.Co.,Ltd. 4275 Carlit Holdings Co.,Ltd. 3553 KYOWA LEATHER CLOTH CO.,LTD. 4295 Faith, Inc. 3649 FINDEX Inc. 4326 INTAGE HOLDINGS Inc. 3660 istyle Inc. 4344 SOURCENEXT CORPORATION 3681 V-cube,Inc. 4671 FALCO HOLDINGS Co.,Ltd. 3751 Japan Asia Group Limited 4779 SOFTBRAIN Co.,Ltd. 3844 COMTURE CORPORATION 4801 CENTRAL SPORTS Co.,LTD. -

Holdings As of June 30, 2021
Units Cost Market Value INTERNATIONAL EQUITY FUND-I International Equities 97.27% International Common Stocks AUSTRALIA ABACUS PROPERTY GROUP 4,781 10,939 11,257 ACCENT GROUP LTD 3,078 2,769 6,447 ADBRI LTD 224,863 495,699 588,197 AFTERPAY LTD 18,765 1,319,481 1,662,401 AGL ENERGY LTD 3,897 48,319 23,926 ALTIUM LTD 11,593 214,343 319,469 ALUMINA LTD 10,311 14,655 12,712 AMP LTD 18,515 29,735 15,687 APA GROUP 2,659 20,218 17,735 APPEN LTD 20,175 310,167 206,065 ARENA REIT 2,151 5,757 5,826 ASX LTD 678 39,359 39,565 ATLAS ARTERIA LTD 5,600 25,917 26,787 AURIZON HOLDINGS LTD 10,404 32,263 29,075 AUSNET SERVICES LTD 9,482 10,386 12,433 AUSTRALIA & NEW ZEALAND BANKIN 22,684 405,150 478,341 AVENTUS GROUP 2,360 4,894 5,580 BANK OF QUEENSLAND LTD 2,738 17,825 18,706 BEACH ENERGY LTD 5,466 6,192 5,108 BEGA CHEESE LTD 1,762 6,992 7,791 BENDIGO & ADELAIDE BANK LTD 2,573 19,560 20,211 BHP GROUP LTD 9,407 243,370 341,584 BHP GROUP PLC 75,164 1,584,327 2,212,544 BLUESCOPE STEEL LTD 2,905 24,121 47,797 BORAL LTD 4,848 16,859 26,679 BRAINCHIP HOLDINGS LTD 5,756 2,588 2,112 BRAMBLES LTD 153,566 1,133,082 1,318,725 BRICKWORKS LTD 375 4,689 7,060 BWP TRUST 2,988 8,177 9,530 CARSALES.COM LTD 466 6,896 6,916 CENTURIA INDUSTRIAL REIT 2,943 6,264 8,191 CENTURIA OFFICE REIT 190,589 261,156 334,222 CHALICE MINING LTD 464 3,129 2,586 CHALLENGER LTD 3,038 15,904 12,335 CHARTER HALL LONG WALE REIT 3,600 12,905 12,793 CHARTER HALL RETAIL REIT 148,478 395,662 422,150 CHARTER HALL SOCIAL INFRASTRUC 2,461 5,340 6,404 CIMIC GROUP LTD 409 6,668 6,072 COCHLEAR LTD 2,492 -

Evaluation of Weissella Cibaria JW15 Probiotic Derived from Fermented
animals Article Evaluation of Weissella Cibaria JW15 Probiotic Derived from Fermented Korean Vegetable Product Supplementation in Diet on Performance Characteristics in Adult Beagle Dog Hao Yang Sun 1, Kun Phil Kim 1, Chun Ho Bae 2, Ae Jin Choi 3, Hyun Dong Paik 4 and In Ho Kim 1,* 1 Department of Animal Resource and Science, Dankook University, Cheonan 31116, Korea 2 Aram Co., Ltd. 54 Gyeongchung-daero 1234 beon-gil, Chowol-eup, Gwangju-si, Gyeonggi-do 12735, Korea 3 National Institute of Agricultural Science, Department of Agro-food Resources, 166 Nongsaengmyeong-ro, Iseo-myeon, Wanju-gun, Jeollabuk-do 55365, Korea 4 Department of Food Science and Biotechnology of Animal Resources, Konkuk University, 120 Neungdong-ro, Gwangjin-gu, Seoul 05029, Korea * Correspondence: [email protected]; Tel.: +82-41-550-3652; Fax: +82-41-559-7881 Received: 19 May 2019; Accepted: 14 August 2019; Published: 20 August 2019 Simple Summary: Dogs are the most popular companion animals worldwide, and their popularity is still increasing. Maintaining health and seeking optimal nutritional feed for dogs is an important component of responsible pet ownership. Probiotics have been widely used in animals; however, little research exists on some probiotic species which have been reported to have good probiotic properties. The present study investigated the effects of Weissella cibaria JW 15 isolated from the traditional Korean fermented vegetable product (kimchi) as a probiotic feed additive on the nutrition and health characteristics in the adult Beagle dogs. The results of this study indicated that Weissella cibaria probiotics have beneficial effects in dogs, which provide evidence and insight for the application of probiotics in dogs. -

CFA Society Taiwan National Taiwan University
CFA Institute Research Challenge Hosted by CFA Society Taiwan National Taiwan University The CFA Institute Research Challenge is a global competition that tests the equity research and valuation, investment report writing, and presentation skills of university students. The following report was prepared in compliance with the Official Rules of the CFA Institute Research Challenge, is submitted by a team of university students as part of this annual educational initiative and should not be considered a professional report. Disclosures: Ownership and material conflicts of interest: The author(s), or a member of their household, of this report does not hold a financial interest in the securities of this company. The author(s), or a member of their household, of this report does not know of the existence of any conflicts of interest that might bias the content or publication of this report. Receipt of compensation: Compensation of the author(s) of this report is not based on investment banking revenue. Position as an officer or director: The author(s), or a member of their household, does not serve as an officer, director or advisory board member of the subject company. Market making: The author(s) does not act as a market maker in the subject company’s securities. Disclaimer: The information set forth herein has been obtained or derived from sources generally available to the public and believed by the author(s) to be reliable, but the author(s) does not make any representation or warranty, express or implied, as to its accuracy or completeness. The information is not intended to be used as the basis of any investment decisions by any person or entity. -

Constituent Changes TOPIX New Index Series (Effective 31 October 2016)
Constituent Changes TOPIX New Index Series (effective 31 October 2016) Published on 7 October 2016 1. Constituents Change (1) TOPIX Core30 Addition( 1 ) Deletion( 1 ) Code Issue Code Issue 6861 KEYENCE CORPORATION 8604 Nomura Holdings, Inc. (2) TOPIX Large70 Addition( 1 ) Deletion( 1 ) Code Issue Code Issue 8604 Nomura Holdings, Inc. 6861 KEYENCE CORPORATION (3) TOPIX Mid400 Addition( 7 ) Deletion( 7 ) Code Issue Code Issue 2201 Morinaga & Co.,Ltd. 1979 Taikisha Ltd. 3938 LINE Corporation 3608 TSI HOLDINGS CO.,LTD. 4043 Tokuyama Corporation 5021 COSMO ENERGY HOLDINGS COMPANY,LIMITED 4095 NIHON PARKERIZING CO.,LTD. 6740 Japan Display Inc. 4587 PeptiDream Inc. 6754 ANRITSU CORPORATION 7458 DAIICHIKOSHO CO.,LTD. 8368 The Hyakugo Bank,Ltd. 8585 Orient Corporation 8544 The Keiyo Bank,Ltd. (4) TOPIX Small Addition(14) Deletion( 7 ) Code Issue Code Issue 1979 Taikisha Ltd. 2201 Morinaga & Co.,Ltd. 3608 TSI HOLDINGS CO.,LTD. 3938 LINE Corporation 5021 COSMO ENERGY HOLDINGS COMPANY,LIMITED 4043 Tokuyama Corporation 6740 Japan Display Inc. 4095 NIHON PARKERIZING CO.,LTD. 6754 ANRITSU CORPORATION 4587 PeptiDream Inc. 8368 The Hyakugo Bank,Ltd. 7458 DAIICHIKOSHO CO.,LTD. 8544 The Keiyo Bank,Ltd. 8585 Orient Corporation * 3221 Yossix Co.,Ltd. * 3445 RS Technologies Co.,Ltd. * 3837 Ad-Sol Nissin Corporation * 3918 PCI Holdings,INC. * 6050 E-Guardian Inc. * 7527 SystemSoft Corporation * 9644 TANABE MANAGEMENT CONSULTING CO.,LTD. * This issue will be newly added to TOPIX new index series after the close of trading on October 28. 2. Number of constituents The Name of TOPIX New Index Current After (As of October 7) (Effective 30 October) TOPIX Core30 30 30 TOPIX Large70 70 70 TOPIX Mid400 400 400 TOPIX Small1 496 500 TOPIX Small2 974 977 Each code means constituents of following index. -

Effect of Weissella Cibaria on Fusobacterium Nucleatum-Induced Interleukin-6 and Interleukin-8 Production in KB Cells
Journal of Bacteriology and Virology 2011. Vol. 41, No. 1 p.9 – 18 DOI 10.4167/jbv.2011.41.1.9 Original Article Effect of Weissella cibaria on Fusobacterium nucleatum-induced Interleukin-6 and Interleukin-8 Production in KB Cells * Mi-Sun Kang1, Hoi-Soon Lim2, Seon-Mi Kim2, Hyun-Chul Lee1 and Jong-Suk Oh1 1Department of Microbiology, School of Medicine, Chonnam National University, Gwangju, 2Dental Science Research Institute, Chonnam National University, Gwangju, Korea Oral microorganisms, including pathogens together with commensals, interact with oral epithelial cells, which can lead to the activation and expression of a variety of inflammatory mediators in epithelial cells. Fusobacterium nucleatum is a filamentous human pathogen that is strongly associated with periodontal diseases. Our previous data suggest that Weissella cibaria, an oral commensal, inhibits the proliferation of periodontopathic bacteria including F. nucleatum. The aim of this study was to examine the effects of W. cibaria on the inflammatory mediators, interleukin (IL)-6 and IL-8, in KB cells stimulated by F. nucleatum. In a reverse transcription-polymerase chain reaction and an enzyme-linked immunosorbent assay, live F. nucleatum alone induced high levels of gene expression and protein release of IL-6 and IL-8, whereas W. cibaria alone did not induce IL-6 and IL-8 responses in KB cells. W. cibaria dose-dependently inhibited the increases of the IL-6 and IL-8 gene expression as well as IL-6 protein level in KB cells which was induced by F. nucleatum. Bacterial viability and its coaggregation with F. nucleatum are not essential in the inhibitory effect of W.