Supplemental Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ACTL6A Promotes the Proliferation of Esophageal Squamous Cell Carcinoma Cells and Correlates with Poor Clinical Outcomes
OncoTargets and Therapy Dovepress open access to scientific and medical research Open Access Full Text Article ORIGINAL RESEARCH ACTL6A Promotes the Proliferation of Esophageal Squamous Cell Carcinoma Cells and Correlates with Poor Clinical Outcomes This article was published in the following Dove Press journal: OncoTargets and Therapy Rui-zhe Li1 Background: ACTL6A, a regulatory subunit of ATP-dependent chromatin-remodeling Yun-yun Li1,2 complexes SWI/SNF, has been identified as a central oncogenic driver in many tumor types. Hui Qin1 Materials and Methods: We used immunohistochemistry (IHC) to detect ACTL6A Shan-shan Li1 expression in esophageal squamous cell carcinoma (ESCC) tissues. Then, the effect of ACTL6A on proliferation and DNA synthesis was explored by using cell counting kit 8 1 Department of Pathology, School of (CCK8) and EdU retention assays. The potential oncogenic mechanism of ACTL6A in Basic Medical Sciences, Zhengzhou University and First Affiliated Hospital of ESCC cells was also analyzed by flow cytometry and Western blotting. We further estab Zhengzhou University, Zhengzhou, lished an ESCC xenograft mouse model to validate the in vitro results. Henan 450000, People’s Republic of China; 2Department of Stomatology, First Results: ACTL6A expression, localized in cancer cell nuclei, was markedly higher in ESCC Affiliated Hospital of Zhengzhou tissues than in the corresponding noncancerous tissues (P<0.001) and was positively asso University, Zhengzhou, Henan 450000, ciated with tumor size, histological differentiation, T stage and tumor-node-metastasis People’s Republic of China (TNM) stage. Kaplan–Meier analysis revealed that high ACTL6A expression was signifi cantly associated with poor overall survival (OS) (P = 0.008, HR= 2.562, 95% CI: 1.241– 5.289), and decision curve analysis (DCA) demonstrated that ACTL6A could increase the clinical prognostic efficiency of the original clinical prediction model. -

Increased ACTL6A Occupancy Within Mswi/SNF Chromatin Remodelers
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.22.435873; this version posted March 22, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Increased ACTL6A Occupancy Within mSWI/SNF Chromatin Remodelers Drives Human Squamous Cell Carcinoma Chiung-Ying Chang1,2,7, Zohar Shipony3,7, Ann Kuo1,2, Kyle M. Loh4,5, William J. Greenleaf3,6, Gerald R. Crabtree1,2,5* 1Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, California 94305, USA. 2Department of Pathology, Stanford University School of Medicine, Stanford, California 94305, USA. 3Department of Genetics, Stanford University School of Medicine, Stanford, California 94305, USA. 4Institute for Stem Cell Biology and Regenerative Medicine, Stanford University School of Medicine, Stanford, California 94305, USA. 5Department of Developmental Biology, Stanford University School of Medicine, Stanford, California 94305, USA. 6Department of Applied Physics, Stanford University, Stanford, California 94305, USA. 7These authors contributed equally. *Corresponding author: Gerald R. Crabtree, [email protected] Summary Mammalian SWI/SNF (BAF) chromatin remodelers play dosage-sensitive roles in many human malignancies and neurologic disorders. The gene encoding the BAF subunit, ACTL6A, is amplified at an early stage in the development of squamous cell carcinomas (SCCs), but its oncogenic role remains unclear. Here we demonstrate that ACTL6A overexpression leads to its stoichiometric assembly into BAF complexes and drives its interaction and engagement with specific regulatory regions in the genome. In normal epithelial cells, ACTL6A was sub-stoichiometric to other BAF subunits. However, increased ACTL6A levels by ectopic expression or in SCC cells led to near-saturation of ACTL6A within BAF complexes. -

Expressed Gene Fusions As Frequent Drivers of Poor Outcomes in Hormone Receptor–Positive Breast Cancer
Published OnlineFirst December 14, 2017; DOI: 10.1158/2159-8290.CD-17-0535 RESEARCH ARTICLE Expressed Gene Fusions as Frequent Drivers of Poor Outcomes in Hormone Receptor–Positive Breast Cancer Karina J. Matissek1,2, Maristela L. Onozato3, Sheng Sun1,2, Zongli Zheng2,3,4, Andrew Schultz1, Jesse Lee3, Kristofer Patel1, Piiha-Lotta Jerevall2,3, Srinivas Vinod Saladi1,2, Allison Macleay3, Mehrad Tavallai1,2, Tanja Badovinac-Crnjevic5, Carlos Barrios6, Nuran Beşe7, Arlene Chan8, Yanin Chavarri-Guerra9, Marcio Debiasi6, Elif Demirdögen10, Ünal Egeli10, Sahsuvar Gökgöz10, Henry Gomez11, Pedro Liedke6, Ismet Tasdelen10, Sahsine Tolunay10, Gustavo Werutsky6, Jessica St. Louis1, Nora Horick12, Dianne M. Finkelstein2,12, Long Phi Le2,3, Aditya Bardia1,2, Paul E. Goss1,2, Dennis C. Sgroi2,3, A. John Iafrate2,3, and Leif W. Ellisen1,2 ABSTRACT We sought to uncover genetic drivers of hormone receptor–positive (HR+) breast cancer, using a targeted next-generation sequencing approach for detecting expressed gene rearrangements without prior knowledge of the fusion partners. We identified inter- genic fusions involving driver genes, including PIK3CA, AKT3, RAF1, and ESR1, in 14% (24/173) of unselected patients with advanced HR+ breast cancer. FISH confirmed the corresponding chromo- somal rearrangements in both primary and metastatic tumors. Expression of novel kinase fusions in nontransformed cells deregulates phosphoprotein signaling, cell proliferation, and survival in three- dimensional culture, whereas expression in HR+ breast cancer models modulates estrogen-dependent growth and confers hormonal therapy resistance in vitro and in vivo. Strikingly, shorter overall survival was observed in patients with rearrangement-positive versus rearrangement-negative tumors. Cor- respondingly, fusions were uncommon (<5%) among 300 patients presenting with primary HR+ breast cancer. -

Inactivation of the PBRM1 Tumor Suppressor Gene Amplifies
Inactivation of the PBRM1 tumor suppressor gene − − amplifies the HIF-response in VHL / clear cell renal carcinoma Wenhua Gaoa, Wei Lib,c, Tengfei Xiaoa,b,c, Xiaole Shirley Liub,c, and William G. Kaelin Jr.a,d,1 aDepartment of Medical Oncology, Dana-Farber Cancer Institute and Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115; bCenter for Functional Cancer Epigenetics, Dana-Farber Cancer Institute, Boston, MA 02215; cDepartment of Biostatistics and Computational Biology, Dana-Farber Cancer Institute and Harvard T.H. Chan School of Public Health, Boston, MA 02115; and dHoward Hughes Medical Institute, Chevy Chase, MD 20815 Contributed by William G. Kaelin, Jr., December 1, 2016 (sent for review October 31, 2016; reviewed by Charles W. M. Roberts and Ali Shilatifard) Most clear cell renal carcinomas (ccRCCs) are initiated by somatic monolayer culture and in soft agar (10). These effects were not, inactivation of the VHL tumor suppressor gene. The VHL gene prod- however, proven to be on-target, and were not interrogated in uct, pVHL, is the substrate recognition unit of an ubiquitin ligase vivo. As a step toward understanding the role of BAF180 in that targets the HIF transcription factor for proteasomal degrada- ccRCC, we asked whether BAF180 participates in the canonical tion; inappropriate expression of HIF target genes drives renal car- PBAF complex in ccRCC cell lines and whether loss of BAF180 cinogenesis. Loss of pVHL is not sufficient, however, to cause ccRCC. measurably alters ccRCC behavior in cell culture and in mice. Additional cooperating genetic events, including intragenic muta- tions and copy number alterations, are required. -

Rats and Axolotls Share a Common Molecular Signature After Spinal Cord Injury Enriched in Collagen-1
Rats and axolotls share a common molecular signature after spinal cord injury enriched in collagen-1 Athanasios Didangelos1, Katalin Bartus1, Jure Tica1, Bernd Roschitzki2, Elizabeth J. Bradbury1 1Wolfson CARD King’s College London, United Kingdom. 2Centre for functional Genomics, ETH Zurich, Switzerland. Running title: spinal cord injury in rats and axolotls Correspondence: A Didangelos: [email protected] SUPPLEMENTAL FIGURES AND LEGENDS Supplemental Fig. 1: Rat 7 days microarray differentially regulated transcripts. A-B: Protein-protein interaction networks of upregulated (A) and downregulated (B) transcripts identified by microarray gene expression profiling of rat SCI (4 sham versus 4 injured spinal cord samples) 7 days post-injury. Microarray expression data and experimental information is publicly available online (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45006) and is also summarised in Supplemental Table 1. Protein-protein interaction networks were performed in StringDB using the full range of protein interaction scores (0.15 – 0.99) to capture maximum evidence of proteins’ interactions. Networks were then further analysed for betweeness centrality and gene ontology (GO) annotations (BinGO) in Cytoscape. Node colour indicates betweeness centrality while edge colour and thickness indicate interaction score based on predicted functional links between nodes (green: low values; red: high values). C-D: The top 10 upregulated (C) or downregulated (D) transcripts sorted by betweeness centrality score in protein-protein interaction networks shown in A & B. E-F: Biological process GO analysis was performed on networks of upregulated and downregulated genes using BinGO in Cytoscape. Graphs indicate the 20 most significant GO categories and the number of genes in each category. -

Supp Table 6.Pdf
Supplementary Table 6. Processes associated to the 2037 SCL candidate target genes ID Symbol Entrez Gene Name Process NM_178114 AMIGO2 adhesion molecule with Ig-like domain 2 adhesion NM_033474 ARVCF armadillo repeat gene deletes in velocardiofacial syndrome adhesion NM_027060 BTBD9 BTB (POZ) domain containing 9 adhesion NM_001039149 CD226 CD226 molecule adhesion NM_010581 CD47 CD47 molecule adhesion NM_023370 CDH23 cadherin-like 23 adhesion NM_207298 CERCAM cerebral endothelial cell adhesion molecule adhesion NM_021719 CLDN15 claudin 15 adhesion NM_009902 CLDN3 claudin 3 adhesion NM_008779 CNTN3 contactin 3 (plasmacytoma associated) adhesion NM_015734 COL5A1 collagen, type V, alpha 1 adhesion NM_007803 CTTN cortactin adhesion NM_009142 CX3CL1 chemokine (C-X3-C motif) ligand 1 adhesion NM_031174 DSCAM Down syndrome cell adhesion molecule adhesion NM_145158 EMILIN2 elastin microfibril interfacer 2 adhesion NM_001081286 FAT1 FAT tumor suppressor homolog 1 (Drosophila) adhesion NM_001080814 FAT3 FAT tumor suppressor homolog 3 (Drosophila) adhesion NM_153795 FERMT3 fermitin family homolog 3 (Drosophila) adhesion NM_010494 ICAM2 intercellular adhesion molecule 2 adhesion NM_023892 ICAM4 (includes EG:3386) intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)adhesion NM_001001979 MEGF10 multiple EGF-like-domains 10 adhesion NM_172522 MEGF11 multiple EGF-like-domains 11 adhesion NM_010739 MUC13 mucin 13, cell surface associated adhesion NM_013610 NINJ1 ninjurin 1 adhesion NM_016718 NINJ2 ninjurin 2 adhesion NM_172932 NLGN3 neuroligin -
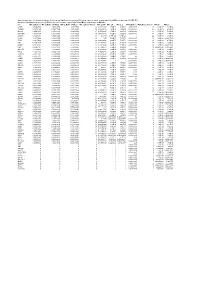
Supplementary Table 6. Summary of Mutation Frequency And
Supplementary table 6. Summary of mutation frequency and significance per microsatellite and per gene in exome sequencing of 24 and MiSeq sequencing of 93 MSI CRCs Summary of mutation frequency and significance per gene in exome sequencing of 24 and MiSeq sequencing of 93 MSI CRCs Gene MiSeq_MutFreq MiSeq_MutFreqCI95High MiSeq_MutFreqCI95Low MiSeq_MutatedTumors MiSeq_NAF MiSeq_P MiSeq_Q WXS_MutFreq WXS_MutatedTumors WXS_P WXS_Q CASP5 0.989247312 1.006838564 0.97165606 92 0.666666667 1.70E-89 4.83E-88 0.791666667 19 2.12E-17 1.33E-14 TGFBR2 0.967741935 0.997877874 0.937605997 90 0.883484163 1.48E-96 6.99E-95 0.833333333 20 4.39E-19 4.13E-16 SLC9A8 0.935483871 0.977386193 0.893581549 87 0.655034325 1.70E-89 4.83E-88 0.458333333 11 5.71E-07 5.25E-05 MIS18BP1 0.913978495 0.961803749 0.86615324 85 0.488888889 7.02E-61 9.07E-60 0.708333333 17 1.51E-08 1.97E-06 ACVR2A 0.892473118 0.945310572 0.839635665 83 0.80625 4.77E-128 6.77E-126 1 24 5.63E-36 2.12E-32 CDH26 0.88172043 0.936801974 0.826638886 82 0.55 4.12E-79 7.31E-78 0.416666667 10 5.46E-06 0.000331959 TFAM 0.88172043 0.936801974 0.826638886 82 0.620192308 4.12E-79 7.31E-78 0.583333333 14 2.52E-10 4.75E-08 ATR 0.860215054 0.919360254 0.801069854 80 0.632996633 2.72E-75 4.30E-74 0.583333333 14 3.89E-09 5.86E-07 AASDH 0.838709677 0.901442635 0.77597672 78 0.832712022 1.27E-71 1.80E-70 0.416666667 10 5.46E-06 0.000331959 USF3 0.806451613 0.873837619 0.739065607 75 0.771428571 5.39E-50 4.25E-49 0.5 12 0.002548628 0.01890901 SLC35F5 0.76344086 0.83592506 0.69095666 71 0.607908352 8.90E-60 1.05E-58 -

Residual Complexes Containing SMARCA2 (BRM) Underlie the Oncogenic Drive of SMARCA4 (BRG1) Mutation
Residual Complexes Containing SMARCA2 (BRM) Underlie the Oncogenic Drive of SMARCA4 (BRG1) Mutation The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Wilson, B. G., K. C. Helming, X. Wang, Y. Kim, F. Vazquez, Z. Jagani, W. C. Hahn, and C. W. M. Roberts. 2014. “Residual Complexes Containing SMARCA2 (BRM) Underlie the Oncogenic Drive of SMARCA4 (BRG1) Mutation.” Molecular and Cellular Biology 34 (6): 1136–44. doi:10.1128/MCB.01372-13. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:41542741 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Residual Complexes Containing SMARCA2 (BRM) Underlie the Oncogenic Drive of SMARCA4 (BRG1) Mutation Boris G. Wilson,a Katherine C. Helming,a Xiaofeng Wang,a Youngha Kim,a Francisca Vazquez,c,d Zainab Jagani,f William C. Hahn,c,d,e Charles W. M. Robertsa,b,d,e Department of Pediatric Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, USAa; Division of Hematology/Oncology, Boston Children’s Hospital, Boston, Massachusetts, USAb; Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, USAc; Cancer Program, Broad Institute of Harvard and Massachusetts Institute of Technology, Boston, Massachusetts, USAd; Center for Cancer Genome Discovery, Dana-Farber Cancer Institute, Boston, Massachusetts, USAe; Novartis Institutes for BioMedical Research, Cambridge, Massachusetts, USAf Collectively, genes encoding subunits of the SWI/SNF (BAF) chromatin remodeling complex are mutated in 20% of all human Downloaded from cancers, with the SMARCA4 (BRG1) subunit being one of the most frequently mutated. -

The Many Roles of BAF (Mswi/SNF) and PBAF Complexes in Cancer
Downloaded from http://perspectivesinmedicine.cshlp.org/ at Cold Spring Harbor Laboratory Library on July 14, 2016 - Published by Cold Spring Harbor Laboratory Press The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer Courtney Hodges, Jacob G. Kirkland, and Gerald R. Crabtree Departments of Pathology, Developmental Biology, and Genetics, Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, California 94305 Correspondence: [email protected] During the last decade, a host of epigenetic mechanisms were found to contribute to cancer and other human diseases. Several genomic studies have revealed that 20% of malignan- cies have alterations of the subunits of polymorphic BRG-/BRM-associated factor (BAF) and Polybromo-associated BAF (PBAF) complexes, making them among the most frequently mutated complexes in cancer. Recurrent mutations arise in genes encoding several BAF/ PBAF subunits, including ARID1A, ARID2, PBRM1, SMARCA4, and SMARCB1. These sub- units share some degree of conservation with subunits from related adenosine triphosphate (ATP)-dependent chromatin remodeling complexes in model organisms, in which a large body of work provides insight into their roles in cancer. Here, we review the roles of BAF- and PBAF-like complexes in these organisms, and relate these findings to recent discoveries in cancer epigenomics. We review several roles of BAF and PBAF complexes in cancer, includ- ing transcriptional regulation, DNA repair, and regulation of chromatin architecture and topology.More recent results highlight the need for new techniques to study these complexes. EPIGENOMICS IN CANCER the full spectrum of achievable cell-type diver- sity for the organism. Because epigenetic regu- roadly defined, epigenetic factors contrib- lation contributes to cell-type functional spe- Bute to the expression state of the genome cialization, it is essential for multicellular life. -

Loss of the Neural-Specific BAF Subunit ACTL6B Relieves PNAS PLUS Repression of Early Response Genes and Causes Recessive Autism
Loss of the neural-specific BAF subunit ACTL6B relieves PNAS PLUS repression of early response genes and causes recessive autism Wendy Wenderskia,b,c,d, Lu Wange,f,g,1, Andrey Krokhotina,b,c,d,1, Jessica J. Walshh, Hongjie Lid,i, Hirotaka Shojij, Shereen Ghoshe,f,g, Renee D. Georgee,f,g, Erik L. Millera,b,c,d, Laura Eliasa,b,c,d, Mark A. Gillespiek, Esther Y. Sona,b,c,d, Brett T. Staahla,b,c,d, Seung Tae Baeke,f,g, Valentina Stanleye,f,g, Cynthia Moncadaa,b,c,d, Zohar Shiponya,b,c,d, Sara B. Linkerl, Maria C. N. Marchettol, Fred H. Gagel, Dillon Chene,f,g, Tipu Sultanm, Maha S. Zakin, Jeffrey A. Ranishk, Tsuyoshi Miyakawaj, Liqun Luod,i, Robert C. Malenkah, Gerald R. Crabtreea,b,c,d,2, and Joseph G. Gleesone,f,g,2 aDepartment of Pathology, Stanford Medical School, Palo Alto, CA 94305; bDepartment of Genetics, Stanford Medical School, Palo Alto, CA 94305; cDepartment of Developmental Biology, Stanford Medical School, Palo Alto, CA 94305; dHoward Hughes Medical Institute, Stanford University, Palo Alto, CA 94305; eDepartment of Neuroscience, University of California San Diego, La Jolla, CA 92037; fHoward Hughes Medical Institute, University of California San Diego, La Jolla, CA 92037; gRady Children’s Institute of Genomic Medicine, University of California San Diego, La Jolla, CA 92037; hNancy Pritztker Laboratory, Department of Psychiatry and Behavioral Sciences, Stanford Medical School, Palo Alto, CA 94305; iDepartment of Biology, Stanford University, Palo Alto, CA 94305; jDivision of Systems Medical Science, Institute for Comprehensive Medical Science, Fujita Health University, 470-1192 Toyoake, Aichi, Japan; kInstitute for Systems Biology, Seattle, WA 98109; lLaboratory of Genetics, The Salk Institute for Biological Studies, La Jolla, CA 92037; mDepartment of Pediatric Neurology, Institute of Child Health, Children Hospital Lahore, 54000 Lahore, Pakistan; and nClinical Genetics Department, Human Genetics and Genome Research Division, National Research Centre, 12311 Cairo, Egypt Edited by Arthur L. -

HNRNPK Maintains Epidermal Progenitor Function Through Transcription of Proliferation Genes and Degrading Differentiation Promoting Mrnas
ARTICLE https://doi.org/10.1038/s41467-019-12238-x OPEN HNRNPK maintains epidermal progenitor function through transcription of proliferation genes and degrading differentiation promoting mRNAs Jingting Li 1, Yifang Chen1, Xiaojun Xu2, Jackson Jones1, Manisha Tiwari1, Ji Ling1, Ying Wang1, Olivier Harismendy 2,3 & George L. Sen1 1234567890():,; Maintenance of high-turnover tissues such as the epidermis requires a balance between stem cell proliferation and differentiation. The molecular mechanisms governing this process are an area of investigation. Here we show that HNRNPK, a multifunctional protein, is necessary to prevent premature differentiation and sustains the proliferative capacity of epidermal stem and progenitor cells. To prevent premature differentiation of progenitor cells, HNRNPK is necessary for DDX6 to bind a subset of mRNAs that code for transcription factors that promote differentiation. Upon binding, these mRNAs such as GRHL3, KLF4, and ZNF750 are degraded through the mRNA degradation pathway, which prevents premature differentiation. To sustain the proliferative capacity of the epidermis, HNRNPK is necessary for RNA Poly- merase II binding to proliferation/self-renewal genes such as MYC, CYR61, FGFBP1, EGFR, and cyclins to promote their expression. Our study establishes a prominent role for HNRNPK in maintaining adult tissue self-renewal through both transcriptional and post-transcriptional mechanisms. 1 Department of Dermatology, Department of Cellular and Molecular Medicine, UCSD Stem Cell Program, University of California, San Diego, La Jolla, CA 92093, USA. 2 Moores Cancer Center, University of California, San Diego, La Jolla, CA 92093, USA. 3 Department of Biomedical Informatics, University of California, San Diego, La Jolla, CA 92093, USA. Correspondence and requests for materials should be addressed to G.L.S. -

ACTL6A Expression Promotes Invasion, Metastasis and Epithelial Mesenchymal Transition of Colon Cancer Zhijun Zeng1,2, Hao Yang1,2 and Shuai Xiao1,2*
Zeng et al. BMC Cancer (2018) 18:1020 https://doi.org/10.1186/s12885-018-4931-3 RESEARCHARTICLE Open Access ACTL6A expression promotes invasion, metastasis and epithelial mesenchymal transition of colon cancer Zhijun Zeng1,2, Hao Yang1,2 and Shuai Xiao1,2* Abstract Background: Metastasisisthemaincauseofdeathinpatientswithadvanced stage colon cancer. Epithelial mesenchymal transition (EMT) plays an important role in invasion and metastasis. Actin-like 6A (ACTL6A) is vital for embryogenesis and differentiation and is also critical for metastasis and EMT in hepatocellular carcinoma, as observed in our previous study. In the present study, we further explored the role of ACTL6A in colon cancer metastasis. Methods: ACTL6A expression levels were analyzed in normal colon, colon adenoma and colon cancer specimens using public databases and tissue samples. ACTL6Aexpressionanditsassociationwithclinicopathologic features of colon cancer patients were also analyzed. ACTL6A-overexpression and ACTL6A-knockdown colon cancer cells were used to perform cytological experiments to explore the potential biological function of ACTL6A in metastasis and EMT in colon cancer. Results: The data from both the Gene Expression Omnibus (GEO) and Oncomine databases showed that ACTL6A expression levels in colon adenoma and cancer were higher than those in normal colon samples. The ACTL6A expression level in fresh colon cancer specimens was also higher than that in the corresponding adjacent normal colon specimens. Patients with high ACTL6A expression directly correlated with advanced pT status, distant metastasis, poor differentiation and microvascular/perineural invasion. ACTL6A overexpression promoted migration and invasion of colon cancer cells, whereas ACTL6A knockdown exhibited the opposite effect in vitro. Moreover, we demonstrated that ACTL6A promoted EMT in colon cancer cells in vitro.