Supplementary Information for Garfinkel Et Al. 2021 SNP In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Dissertation Entitled Uncovering Cannabinoid Signaling in C. Elegans
A Dissertation Entitled Uncovering Cannabinoid Signaling in C. elegans: A New Platform to Study the Effects of Medicinal Cannabis By Mitchell Duane Oakes Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Doctor of Philosophy Degree in Biology ________________________________________ Dr. Richard Komuniecki, Committee Chair _______________________________________ Dr. Bruce Bamber, Committee Member ________________________________________ Dr. Patricia Komuniecki, Committee Member ________________________________________ Dr. Robert Steven, Committee Member ________________________________________ Dr. Ajith Karunarathne, Committee Member ________________________________________ Dr. Jianyang Du, Committee Member ________________________________________ Dr. Amanda Bryant-Friedrich, Dean College of Graduate Studies The University of Toledo August 2018 Copyright 2018, Mitchell Duane Oakes This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. An Abstract of Uncovering Cannabinoid Signaling in C. elegans: A New Platform to Study the Effects of Medical Cannabis By Mitchell Duane Oakes Submitted to the Graduate Faculty as partial fulfillment of the requirements for the Doctor of Philosophy Degree in Biology The University of Toledo August 2018 Cannabis or marijuana, a popular recreational drug, alters sensory perception and exerts a range of medicinal benefits. The present study demonstrates that C. elegans exposed to -

CBD (Cannabidiol)
TRANSPORTATION RESEARCH BOARD Driving Toward the Truth - Dispelling the Myths About Cannabis Products February 10, 2021 @NASEMTRB #TRBwebinar PDH Certification The Transportation Research Board has met the standards and Information: requirements of the Registered Continuing Education Providers •1.5 Professional Development Program. Credit earned on completion Hour (PDH) – see follow-up of this program will be reported to email for instructions RCEP. A certificate of completion will •You must attend the entire be issued to participants that have registered and attended the entire webinar to be eligible to receive session. As such, it does not include PDH credits content that may be deemed or •Questions? Contact Reggie construed to be an approval or Gillum at [email protected] endorsement by RCEP. #TRBwebinar Learning Objectives 1. Identify impacts of the Farm Bill on use of THC and CBD products 2. Describe the toxicology of THC and CBD products 3. Discuss how THC and CBD products affect driving performance and crash risk #TRBwebinar TRB Standing Committee on Impairment in Transportation (ACS50) TRB Webinar: Driving Toward the Truth - Dispelling the Myths About Cannabis Products Dr. Barry K. Logan Executive Director, Center for Forensic Science Research and Education (CFSRE); Senior Vice President of Forensic Sciences, and Chief Scientist at NMS Labs Michelle Peace, Ph.D. Associate Professor and PI, Laboratory for Forensic Toxicology Research Department of Forensic Science, Virginia Commonwealth University Dr. Darrin Grondel Vice President, -

Endocannabinoid System Dysregulation from Acetaminophen Use May Lead to Autism Spectrum Disorder: Could Cannabinoid Treatment Be Efficacious?
molecules Review Endocannabinoid System Dysregulation from Acetaminophen Use May Lead to Autism Spectrum Disorder: Could Cannabinoid Treatment Be Efficacious? Stephen Schultz 1, Georgianna G. Gould 1, Nicola Antonucci 2, Anna Lisa Brigida 3 and Dario Siniscalco 4,* 1 Department of Cellular and Integrative Physiology, Center for Biomedical Neuroscience, University of Texas (UT) Health Science Center San Antonio, San Antonio, TX 78229, USA; [email protected] (S.S.); [email protected] (G.G.G.) 2 Biomedical Centre for Autism Research and Therapy, 70126 Bari, Italy; [email protected] 3 Department of Precision Medicine, University of Campania, 80138 Naples, Italy; [email protected] 4 Department of Experimental Medicine, University of Campania, 80138 Naples, Italy * Correspondence: [email protected] Abstract: Persistent deficits in social communication and interaction, and restricted, repetitive pat- terns of behavior, interests or activities, are the core items characterizing autism spectrum disorder (ASD). Strong inflammation states have been reported to be associated with ASD. The endocannabi- noid system (ECS) may be involved in ASD pathophysiology. This complex network of lipid signal- ing pathways comprises arachidonic acid and 2-arachidonoyl glycerol-derived compounds, their G-protein-coupled receptors (cannabinoid receptors CB1 and CB2) and the associated enzymes. Alter- Citation: Schultz, S.; Gould, G.G.; ations of the ECS have been reported in both the brain and the immune system of ASD subjects. ASD Antonucci, N.; Brigida, A.L.; Siniscalco, D. Endocannabinoid children show low EC tone as indicated by low blood levels of endocannabinoids. Acetaminophen System Dysregulation from use has been reported to be associated with an increased risk of ASD. -

Cannabinoids: Novel Molecules with Significant Clinical Utility
CANNABINOIDS: NOVEL MOLECULES WITH SIGNIFICANT CLINICAL UTILITY NOEL ROBERT WILLIAMS MD FACOG DIRECTOR OPTIMAL HEALTH ASSOCIATES OKLAHOMA CITY, OKLAHOMA How Did We Get Here? • In November 2012 Tikun Olam, an Israeli medical cannabis facility, announced a new strain of the plant which has only CBD as an active ingredient, and virtually no THC, providing some of the medicinal benefits of cannabis without euphoria. The Researchers said the cannabis plant, enriched with CBD, “can be used for treating diseases like rheumatoid arthritis, colitis, liver inflammation, heart disease and diabetes.” Cannabis CBD like in this article is legally derived from the hemp plant. • CBD is the major non-psychoactive component of Cannabis Sativa (Hemp). Hemp plants are selectively developed and grown to contain high amounts of CBD and very low amounts of the psychoactive component THC found in marijuana. A few CBD oil manufacturers further purify their products to contain high amounts of CBD and no THC. 2014 Farm Bill Terminology • Active Ingredient • Zero THC • Cannabidiol • Isolate • PCR – Phytocannabinoid-Rich • Hemp Oil Extract • “Recommendation” vs. “Prescribed” • Full Spectrum Endocannabinoids (AEA) Phytocannabinoids Full Spectrum & Active Ingredient • CBD – Cannabidiol • A major phytocannabinoid, accounting for as much as 85% of the plant’s extract • CBC – Cannabichromene • Anti-inflammatory & anti-fungal effects have been seen • CBG – Cannabigerol • The parent molecule from which many other cannabinoids are made • CBDV – Cannabidivarin • A homolog of CBD that has been reported to have powerful anti-convulsive effects • CBN – Cannabinol • Sleep & Appetite regulation • Terpenes • Wide spectrum of non-psychoactive molecules that are know to act on neural receptors and neurotransmitters, enhance norepinephrine activity, and potentially increases dopamine activity. -

Pharmacodynamics of Cannabinoids
Open Access Archives of Pharmacy and Pharmaceutical Sciences Review Article Pharmacodynamics of cannabinoids Alexandra Sulcova* ISSN ICCI - International Cannabis and Cannabinoids Institute, Jachymova 26/2, 110 00 Praha, 2639-992X Czech Republic “Pharmacodynamics of cannabinoids “(i.e. a set of biological effects elicited in the *Address for Correspondence: Alexandra Sulcova, M.D, Ph.D, Professor of Pharmacology, living organism by interaction with its biochemical and biophysical functions up to the FCMA, FECNP, FCINP, ICCI - International cellular level) is studied for a long time during both, physiological and pathological Cannabis and Cannabinoids Institute, Jachymova conditions. Cannabinoids received their names according to their natural occurrence 26/2, 110 00 Praha, Czech Republic, Tel: 420 732167678; Email: [email protected] as constituents of Cannabis sativa L. (marijuana). The species was classiied in the “Linnaeus’s Species Plantarum (1753)”, the word “sativa” means things that are Submitted: 12 April 2019 Approved: 07 May 2019 cultivated [1]. For ages, people have used cannabis-based preparations for healing and Published: 08 May 2019 pain suppression until the discovery (in 1897) of aspirin (acetylsalicylic acid) which contemporary medicine uses until today. Chemical investigation of marijuana conirmed Copyright: © 2019 Sulcova A. This is an open access article distributed under the Creative various cannabinoid-type components called cannabinoids (presently estimated at Commons Attribution License, which permits about 150). Regarding their possible pharmacodynamic effects, tetrahydrocannabinol unrestricted use, distribution, and reproduction (THC) and cannabidiol (CBD) are the most explored. The determination of THC structure in any medium, provided the original work is properly cited by means of nuclear magnetic resonance imaging increased sharply the number of professional scientiic reports dealing with the studies of THC pharmacodynamic mechanisms of action [2]. -

On Brain Excitation and Inhibition Systems in Adults with and Without Autism Spectrum Disorder
Pretzsch et al. Translational Psychiatry (2019) 9:313 https://doi.org/10.1038/s41398-019-0654-8 Translational Psychiatry ARTICLE Open Access Effects of cannabidivarin (CBDV) on brain excitation and inhibition systems in adults with and without Autism Spectrum Disorder (ASD): a single dose trial during magnetic resonance spectroscopy Charlotte M Pretzsch1, Bogdan Voinescu 1,DavidLythgoe 2, Jamie Horder1, Maria Andreina Mendez1, Robert Wichers 1, Laura Ajram1,GlynisIvin3, Martin Heasman3, Richard A. E. Edden4, Steven Williams 2, Declan G. M. Murphy 1, Eileen Daly1 and Gráinne M. McAlonan1 Abstract Autism spectrum disorder (ASD) is a high cost neurodevelopmental condition; and there are currently no effective pharmacological treatments for its core symptoms. This has led some families and researchers to trial alternative remedies – including the non-intoxicating Cannabis sativa-derived compound cannabidivarin (CBDV). However, how CBDV affects the human brain is unknown. Previous (pre)clinical evidence suggests that CBDV may modulate brain excitatory-inhibitory systems, which are implicated in ASD. Hence, our main aim was to test, for the first time, if CBDV shifts glutamate and/or GABA metabolites – markers of the brain’s primary excitatory and inhibitory system - in both the ‘typical’ and autistic brain. Our subsidiary aim was to determine whether, within ASD, brain responsivity to CBDV challenge is related to baseline biological phenotype. We tested this using a repeated-measures, double-blind, 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; randomized-order, cross-over design. We used magnetic resonance spectroscopy (MRS) to compare glutamate (Glx = glutamate + glutamine) and GABA + (GABA + macromolecules) levels following placebo (baseline) and 600 mg CBDV in 34 healthy men with (n = 17) and without (n = 17) ASD. -
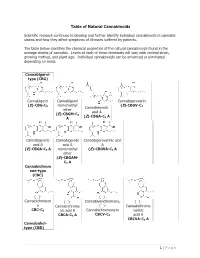
Table of Natural Cannabinoids
Table of Natural Cannabinoids Scientific research continues to develop and further identify individual cannabinoids in cannabis strains and how they affect symptoms of illnesses suffered by patients. The table below identifies the chemical properties of the natural cannabinoids found in the average strains of cannabis. Levels of each of these chemicals will vary with varietal strain, growing method, and plant age. Individual cannabinoids can be enhanced or eliminated depending on need. Cannabigerol- type (CBG) Cannabigerol Cannabigerol Cannabigerovarin (E)-CBG-C monomethyl (E)-CBGV-C 5 Cannabinerolic 3 ether acid A (E)-CBGM-C 5 (Z)-CBGA-C A A 5 Cannabigerolic Cannabigerolic Cannabigerovarinic acid acid A acid A A (E)-CBGA-C5 A monomethyl (E)-CBGVA-C3 A ether (E)-CBGAM- C5 A Cannabichrom ene-type (CBC) (±)- (±)- Cannabichromen (±)- Cannabivarichromene, (±)- e Cannabichrome (±)- Cannabichrome CBC-C5 nic acid A Cannabichromevarin varinic CBCA-C5 A CBCV-C3 acid A CBCVA-C3 A Cannabidiol- type (CBD) 1 | Page (−)-Cannabidiol Cannabidiol Cannabidiol-C4 (−)- Cannabidiorc CBD-C5 momomethyl CBD-C4 Cannabidivarin ol ether CBDV-C3 CBD-C1 CBDM-C5 Cannabidiolic Cannabidivarini acid c acid CBDA-C5 CBDVA-C3 Cannabinodiol- type (CBND) Cannabinodiol Cannabinodivar CBND-C5 in CBND-C3 Tetrahydrocan nabinol-type (THC) 9 9 9 Δ - Δ - Δ - Δ9- Tetrahydrocanna Tetrahydrocan Tetrahydrocannabivarin 9 Tetrahydrocan binol nabinol-C4 Δ -THCV-C3 9 9 nabiorcol Δ -THC-C5 Δ -THC-C4 9 Δ -THCO-C1 9 9 Δ -Tetrahydro- Δ9-Tetrahydro- Δ -Tetrahydro- Δ9-Tetrahydro- cannabinolic -

Cannabinoids for People with ASD: a Systematic 3 Review of Published and Ongoing Studies
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 17 July 2020 doi:10.20944/preprints202007.0373.v1 Peer-reviewed version available at Brain Sci. 2020, 10, 572; doi:10.3390/brainsci10090572 1 Review 2 Cannabinoids for People with ASD: A Systematic 3 Review of Published and Ongoing Studies 4 Laura Fusar-Poli1, Vito Cavone1 , Silvia Tinacci1, Ilaria Concas1, Antonino Petralia1, Maria 5 Salvina Signorelli1, Covadonga M. Díaz-Caneja2, Eugenio Aguglia1* 6 1 Department of Clinical and Experimental Medicine, Psychiatry Unit, University of Catania, via Santa Sofia 7 78, 95123, Catania, Italy; [email protected] (L.F.-P.); [email protected] (V.C.); 8 [email protected] (S.T.); [email protected] (I.C.); [email protected] (A.P.); 9 [email protected] (M.S.S.) 10 2 Department of Child and Adolescent Psychiatry, Hospital General Universitario Gregorio Marañón, 11 Instituto de Investigación Sanitaria Gregorio Marañón (IiSGM), School of Medicine, Universidad 12 Complutense, Centro de Investigación Biomédica en Red del área de Salud Mental (CIBERSAM), Calle Ibiza, 13 43, 28009, Madrid, Spain; [email protected] (C.M.D.-C.) 14 15 *Correspondence: [email protected]; Tel.: +39 095 378 2470 (E.A.) 16 Abstract: The etiopathogenesis of autism spectrum disorder (ASD) remains largely unclear. Among 17 other biological hypotheses, researchers have evidenced an imbalance in the endocannabinoid (eCB) 18 system, which regulates some functions typically impaired in ASD, such as emotional responses 19 and social interaction. Also, cannabidiol (CBD), the non-intoxicating component of Cannabis sativa, 20 has been recently approved for treatment-resistant epilepsy. Seizures represent frequent medical 21 comorbidities of ASD and could be responsible for the onset or worsening of behavioral problems. -

Study of the Effects of the Cannabinoids Anandamide and Cannabidiol on the Feeding Processes of Tetrahymena Pyriformis
Study of the Effects of the Cannabinoids Anandamide and Cannabidiol on the feeding processes of Tetrahymena pyriformis Charou Jaisswar Student ID: 32171241 December 2019 Masters by Research Project Thesis Supervised by Dr Jackie Parry and Dr Karen L. Wright Declaration This thesis is entirely my own work and has not been submitted in full or in part for the award of a higher degree at any other educational institution. No sections of this thesis have been published. Word count: 23,091 1 Table of Contents Abstract 4 Glossary 5 1. Introduction 8 1.1 The human endocannabinoid system 8 1.2 Cannabinoids 8 1.2.1. Phytocannabinoids 8 1.2.2. Endogenous cannabinoids (Endocannabinoids) 9 1.3 Biosynthetic and degradation enzymes 10 1.4 Endocannabinoid receptors and cannabinoid interactions 13 1.4.1. CB1 13 1.4.2. CB2 15 1.4.3. TRPV1 15 1.4.4. Other receptors 16 1.5. Single celled protists and cannabinoids 17 1.5.1. Introduction to protists 17 1.5.2. Presence of ECS components in ciliates 18 1.5.2.1. Endocannabinoids 18 1.5.2.2. Enzymes 18 1.5.2.3. Receptors 19 1.5.3. Effect of exogenous cannabinoids on protists 19 1.5.4. Ciliate feeding process 20 1.5.4.1. Ingestion 20 1.5.4.2. Digestion 22 1.6. Rationale and aims of this study 24 2. Materials and Methods 26 2.1. Organisms and maintenance 26 2.1.1. Preparation and maintenance of prey 26 2.1.2. Preparation and maintenance of Tetrahymena pyriformis 26 2.2. -
![) (51) International Patent Classification: (71) Applicant: GENOMATICA, INC. [US/US]; 4757 Nexus C02F 1/38 (2006.0 1) (72) Inven](https://docslib.b-cdn.net/cover/2660/51-international-patent-classification-71-applicant-genomatica-inc-us-us-4757-nexus-c02f-1-38-2006-0-1-72-inven-2932660.webp)
) (51) International Patent Classification: (71) Applicant: GENOMATICA, INC. [US/US]; 4757 Nexus C02F 1/38 (2006.0 1) (72) Inven
) ( (51) International Patent Classification: (71) Applicant: GENOMATICA, INC. [US/US]; 4757 Nexus B 11) 17/038 (2006.0 1) C02F 1/38 (2006.0 1) Center Drive, San Diego, California 92121 (US). B 11) 17/025 (2006.01) (72) Inventors: SUOMINEN, Lauri, H.; 4757 Nexus Center (21) International Application Number: Drive, San Diego, California 92121 (US). LIEVENSE, PCT/US2020/0 15904 Jefferey; 4757 Nexus Center Drive, San Diego, Califor¬ nia 92121 (US). LIU, Aiguo; 4757 Nexus Center Drive, (22) International Filing Date: SanDiego, California 92121 (US). MARTINEZ-GONZA- 30 January 2020 (30.01.2020) LEZ, Jorge, Jesus; 4757 Nexus Center Drive, San Diego, (25) Filing Language: English California 92 121 (US). KO, Myong Kon; 4757 Nexus Cen¬ ter Drive, San Diego, California 92121 (US). NAN, Wei; (26) Publication Language: English 4757 Nexus Center Drive, San Diego, California 92121 (30) Priority Data: (US). 62/798,909 30 January 2019 (30.01.2019) US (74) Agent: WEAVER, Paul, L. et al.; Kagan Binder, PLLC, 62/802,101 06 February 2019 (06.02.2019) US Suite 200, Maple Island Building, 22 1 Main Street North, 62/934,371 12 November 2019 (12. 11.2019) US Stillwater, Minnesota 55082 (US). (54) Title: RECOVERY, DECARBOXYLATION, AND PURIFICATION OF CANNABINOIDS FROM ENGINEERED CELL CULTURES (57) Abstract: Methods of recovering cannabinoids from cell cultures include methods comprising steps of separating the cell culture at a temperature above the melting point of the cannabinoid to separate a light phase comprising liquid state cannabinoid from a heavy phase; and methods comprising treating the cell culture at a temperature below the melting point of the cannabinoid to separate a light phase from a heavy phase comprising solid state cannabinoid. -

Cannabigerol Is a Potential Therapeutic Agent in a Novel Combined Therapy for Glioblastoma
Supplemental material Cannabigerol is a potential therapeutic agent in a novel combined therapy for glioblastoma Tamara T. Lah, Metka Novak, Milagros A. Pena Almidon, Oliviero Marinelli, Barbara Žvar Baškovič, Bernarda Majc, Mateja Mlinar, Roman Bošnjak, Barbara Breznik, Roby Zomer, and Massimo Nabissi Figure S1. Biosynthesis and metabolism of cannabigerol (CBG). The enzyme prenyltransferase catalyses the conversion of olivetolic acid into CBG in the cannabis plant. CBG is the intermediate biosynthetic precursor of delta‐9‐tetrahydrocannabinol (THC)‐acid and cannabidiol (CBD)‐acid and is converted to THC, CBD, and cannabichromene (CBC), which is then converted into THC, CBD, or CBG by specific synthases (adapted from [30]). Cells 2021, 10, 340. https://doi.org/10.3390/cells10020340 www.mdpi.com/journal/cells Cells 2021, 10, 340 2 of 3 Figure S2. Cell death determination by flow cytometry using Annexin V‐FITC and propidium iodide staining. Cells (U87 and NCH44) were treated with IC50 concentrations of compounds CBG, CBD, and THC for 48 h. Cells were labelled with Annexin V‐FITC and propidium iodide and analysed by flow cytometry. The dot blots represent the results from three biological repeats. Table S1: High‐performance liquid chromatography purity results for the CBG, CBD, and THC solutions used in this study. Cannabinoids Cannabinoids Cannabinoids Concentration Concentration Concentration in CBG in CBD in CBD (mg/ml) (mg/ml) (mg/ml) solution solution solution CBDVA BDL* CBDVA BDL* CBDVA BDL* CBDV BDL* CBDV BDL* CBDV BDL* CBDA BDL* -

Phytocannabinoids
Phytocannabinoids More than 100 structurally and physiologically distinct cannabinoid compounds are unique to plants of the genus Cannabis, known collectively as phytocannabinoids. Cayman Chemical offers authentic reference standards for the most prominent of these phytocannabinoids, as well as their metabolites, from our ISO/IEC 17025 and ISO 17034 certified labs. We are working diligently to introduce many more notable phytocannabinoids. If you cannot find a particular compound of interest in our catalog, please contact us for a custom synthesis estimate. Primary Active Constituents Item No. Product Name Metabolites ISO60156 Cannabidiol (CRM)* Item No. Product Name ISO60158 Δ8-THC (CRM)* 21667 (±)-11-hydroxy-Δ9-THC (CRM)* ISO60157 Δ9-THC (CRM)* 20754 (±)-11-nor-9-carboxy-Δ9-THC (CRM)* *Isotopically labeled standard available *Isotopically labeled standard available Naturally Occurring Acids THC Isomers Item No. Product Name Item No. Product Name 9001573 (±)-Cannabichromenic Acid (dicyclohexylamine salt) 25707 (6aR,9S)-Δ10-THC 18090 Cannabidiolic Acid (CRM) 26528 9(R)-Δ6a,10a-THC 20019 Cannabigerolic Acid (CRM) 26529 9(S)-Δ6a,10a-THC ISO60175 THCA-A (CRM) 26283 Varinolic Acid Discover Our Multi-Component Varinol Series Phytocannabinoid CRM Mixtures Item No. Product Name Available to quantify up to 10 21974 (±)-Cannabichromevarin (CRM) prevalent phytocannabinoids 20165 Cannabidivarin (CRM) Learn more 21664 Cannabivarin on Page 2 18091 Tetrahydrocannabivarin (CRM) of this Other Cannabinoids of Interest brochure Item No. Product Name ISO60163