Optimization and Application of Loop-Mediated Isothermal Amplifcation Detection Method on Little Cherry Virus 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Establishing 15 IP Tribunals Nationwide, Chinese Courts Further Concentrate Jurisdiction Over IP Matters
Establishing 15 IP Tribunals Nationwide, Chinese Courts Further Concentrate Jurisdiction Over IP Matters March 15, 2018 Patent and ITC Litigation China has continued to develop its adjudicatory framework for intellectual property disputes with the establishment of three Intellectual Property Tribunals (“IP Tribunals”) this month. This reform began with the establishment of three specialized IP Courts in Beijing, Shanghai, and Guangzhou at the end of 2014, and has been furthered with the establishment of IP Tribunals in 10 provinces and two cities/municipalities around the country. For companies facing an IP dispute in China, understanding this framework in order to select the appropriate jurisdiction for a case can have a significant impact on the time to resolution, as well as the ultimate merits of the case. Most significantly, through the establishment of these IP Tribunals many Chinese courts have been stripped of their jurisdiction over IP matters in favor of the IP Tribunals. This has led to a fundamental change to the forum selection strategies of both multinational and Chinese companies. The three IP Tribunals established on the first two days of March 2018 are located in Tianjin Municipality, and cities of Changsha and Zhengzhou respectively. This brings the number of IP Tribunals that have been set up across 10 provinces and two cities/municipalities in China since January 2017 to a total of 15. The most unique aspect of the specialized IP Tribunals is that they have cross-regional1 and exclusive jurisdiction over IP matters in significant first-instance2 cases (i.e., those generally including disputes involving patents, new varieties of plants, integrated circuit layout and design, technical-related trade secrets, software, the recognition of well-known trademarks, and other IP cases in which the damages sought exceed a certain amount)3. -

A Case Study of the Weishan REE Deposit, China
minerals Article REE Enrichment during Magmatic–Hydrothermal Processes in Carbonatite-Related REE Deposits: A Case Study of the Weishan REE Deposit, China Yu-heng Jia 1,2 and Yan Liu 2,3,* 1 Earth Science Institute, Guilin University of Technology, Guilin 541004, China; [email protected] 2 Key Laboratory of Deep-Earth Dynamics of Ministry of Natural Resources, Institute of Geology, Chinese Academy of Geological Science, Beijing 100037, China 3 Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou 511458, China * Correspondence: [email protected] Received: 22 August 2019; Accepted: 20 December 2019; Published: 27 December 2019 Abstract: The Weishan carbonatite-related rare earth element (REE) deposit in China contains both high- and low-grade REE mineralization and is an informative case study for the investigation of magmatic–hydrothermal REE enrichment processes in such deposits. The main REE-bearing mineral is bastnäsite, with lesser parisite and monazite. REE mineralization occurred at a late stage of hydrothermal evolution and was followed by a sulfide stage. Barite, calcite, and strontianite appear homogeneous in back-scattered electron images and have high REE contents of 103–217, 146–13,120, and 194–16,412 ppm in their mineral lattices, respectively. Two enrichment processes were necessary for the formation of the Weishan deposit: Production of mineralized carbonatite and subsequent enrichment by magmatic–hydrothermal processes. The geological setting and petrographic characteristics of the Weishan deposit indicate that two main factors facilitated REE enrichment: (1) fractures that facilitated circulation of ore-forming fluids and provided space for REE precipitation and (2) high ore fluorite 2 and barite contents resulting in high F− and SO4 − concentrations in the ore-forming fluids that promoted REE transport and deposition. -

A New Indicator to Assess Public Perception of Air Pollution Based on Complaint Data
applied sciences Article A New Indicator to Assess Public Perception of Air Pollution Based on Complaint Data Yong Sun 1 , Fengxiang Jin 2, Yan Zheng 1, Min Ji 1,* and Huimeng Wang 2,3 1 College of Geodesy and Geomatics, Shandong University of Science and Technology, Qingdao 266590, China; [email protected] (Y.S.); [email protected] (Y.Z.) 2 College of Surveying and Geo-Informatics, Shandong Jianzhu University, Ji’nan 250101, China; [email protected] (F.J.); [email protected] (H.W.) 3 State Key Laboratory of Resources and Environmental Information System, Institute of Geographic Science and Natural Resources Research, Chinese Academy of Sciences, Beijing 100101, China * Correspondence: [email protected] Abstract: Severe air pollution problems have led to a rise in the Chinese public’s concern, and it is necessary to use monitoring stations to monitor and evaluate pollutant levels. However, monitoring stations are limited, and the public is everywhere. It is also essential to understand the public’s awareness and behavioral response to air pollution. Air pollution complaint data can more directly reflect the public’s real air quality perception than social media data. Therefore, based on air pollution complaint data and sentiment analysis, we proposed a new air pollution perception index (APPI) in this paper. Firstly, we constructed the emotional dictionary for air pollution and used sentiment analysis to calculate public complaints’ emotional intensity. Secondly, we used the piecewise function to obtain the APPI based on the complaint Kernel density and complaint emotion Kriging interpolation, and we further analyzed the change of center of gravity of the APPI. -

Annual Report Annual Report 2020
2020 Annual Report Annual Report 2020 For further details about information disclosure, please visit the website of Yanzhou Coal Mining Company Limited at Important Notice The Board, Supervisory Committee and the Directors, Supervisors and senior management of the Company warrant the authenticity, accuracy and completeness of the information contained in the annual report and there are no misrepresentations, misleading statements contained in or material omissions from the annual report for which they shall assume joint and several responsibilities. The 2020 Annual Report of Yanzhou Coal Mining Company Limited has been approved by the eleventh meeting of the eighth session of the Board. All ten Directors of quorum attended the meeting. SHINEWING (HK) CPA Limited issued the standard independent auditor report with clean opinion for the Company. Mr. Li Xiyong, Chairman of the Board, Mr. Zhao Qingchun, Chief Financial Officer, and Mr. Xu Jian, head of Finance Management Department, hereby warrant the authenticity, accuracy and completeness of the financial statements contained in this annual report. The Board of the Company proposed to distribute a cash dividend of RMB10.00 per ten shares (tax inclusive) for the year of 2020 based on the number of shares on the record date of the dividend and equity distribution. The forward-looking statements contained in this annual report regarding the Company’s future plans do not constitute any substantive commitment to investors and investors are reminded of the investment risks. There was no appropriation of funds of the Company by the Controlling Shareholder or its related parties for non-operational activities. There were no guarantees granted to external parties by the Company without complying with the prescribed decision-making procedures. -

Reliability Optimization of a Railway Network
sustainability Article Reliability Optimization of a Railway Network Xuelei Meng 1,2,*, Yahui Wang 3, Limin Jia 2 and Lei Li 4 1 School of Traffic and Transportation, Lanzhou Jiaotong University, Lanzhou 730070, Gansu, China 2 State Key Laboratory of Rail Traffic Control and Safety, Beijing Jiaotong University, Beijing 100044, China; [email protected] 3 School of Foreign Languages, Lanzhou Jiaotong University, Lanzhou 730070, Gansu, China; [email protected] 4 Key Laboratory of Urban Rail Transit Intelligent Operation and Maintenance Technology & Equipment of Zhejiang Province, Zhejiang Normal University, Jinhua 321004, Zhejiang, China; [email protected] * Correspondence: [email protected] Received: 7 October 2020; Accepted: 14 November 2020; Published: 24 November 2020 Abstract: With the increase of the railway operating mileage, the railway network is becoming more and more complicated. We expect to build more railway lines to offer the possibility to offer more high quality service for the passengers, while the investment is often limited. Therefore, it is very important to decide the pairs of cities to add new railway lines under the condition of limited construction investment in order to optimize the railway line network to maximize the reliability of the railway network to deal with the railway passenger transport task under emergency conditions. In this paper, we firstly define the reliability of the railway networks based on probability theory by analyzing three minor cases. Then we construct a reliability optimization model for the railway network to solve the problem, expecting to enhance the railway network with the limited investment. The goal is to make an optimal decision when choosing where to add new railway lines to maximize the reliability of the whole railway network, taking the construction investment as the main constraint, which is turned to the building mileage limit. -

Appendix A. the Structural PVAR Model the Structural PVAR Model Analyzing the Dynamics Among Structure (S), Pollution (P), Income (E) and Health (H) Is
Appendix A. The structural PVAR model The structural PVAR model analyzing the dynamics among structure (S), pollution (P), income (E) and health (H) is , , , , ,11 ,12 ,13 ,14 = − + + (A.1) , , , , ⎛ 21 22 23 24⎞ − ⎛ ⎞ ⎛ ⎞ =1 � � ∑ , , , , � − � ⎜ 31 32 33 34⎟ ⎜ ⎟ ⎜⎟ − 41 42 43 44 Define = ( ,⎝ , , ) , using matrix,⎠ Eq. (A.1) can⎝ be⎠ rewritten⎝ ⎠ as = ′ + + (A.2) The 4×4 matrix = ∑= 1reflects− the contemporaneous relation of the �� variables in , and = , are the coefficients of the lagged endogenous variables. Because both contemporaneous � � and long-run relationships are accounted for, we assume that the structural shock is homoskedastic, that is ~( , ), = ⎛ ⎞ ⎜ ⎟ ⎝ ⎠ To estimate eq. (A.2), transform Eq. (A.1) into the reduced form = + + , ~( , ) (A.3) where ∑=1 − = , = , = . Since matrix A is of full −rank, the covariance− matrix− Ω is no longer diagonal. In fact, we have = ( ) (A.4) To recover the structural− parameters− ′ in and , we must impose additional restrictions a priori. Note that the left-hand side of Eq. (A.4) contains 10 coefficients, because is symmetric. But on the right-hand side of (A.2), the number of unknowns in and are 16 and 4, respectively. Therefore, to pin down and , we need 16 + 4 − 10 = 10 more restrictions. Applying Cholesky decomposition, we impose a lower triangular structure on 0 0 0 0 0 = 11 0 21 22 � 31 32 � 33 To finally identify and , we41 should42 either set = = = = 1, 43 44 or = = = = 1 . Based on these restrictions, the PVAR model is 11 22 33 44 transformed into a recursive system. To see the impacts of shocks, we transform Eq. (A.1) to = + (A.5) 1 = � − ∑ � where L is the lag operator. -

Supply Chain Strategic Alliances Partner Selection for Rizhao Port
International Journal of Scientific and Research Publications, Volume 4, Issue 11, November 2014 1 ISSN 2250-3153 Supply Chain Strategic Alliances Partner Selection for Rizhao Port Libin Guo School of Management, Qufu Normal University,Rizhao276826,China Abstract- Under the background of port supply chain strategic alliances,we clarify the current situation of Rizhao Port by SWOT analysis and find out the ST strategy for it, furthermore, we screen out the optimal cooperation partner for Rizhao Port under different alliance forms range from horizontal integration and vertical integration to the blended dynamic logistics alliance. Index Terms- Port Supply Chain; the SWOT Analysis; RiZhao Port; Strategical alliance I. INTRODUCTION ort supply chain, a main constituent of port logistics, includes the levels such as informationization, automation and networking, P and emphasizes the modern management of material transportation chain of each link and the extension of comprehensive services.Meanwhile, due to the connection between ports and suppliers and consumers all over the world through shippping companies and land forwarding agents, a port supply chain integrating many means1 of transportation and types of logistics is formed, and it becomes a relatively best main part and link for the coordinated management of supply chain. Therefore, we can start from the analysis of the SWOT of Rizhao port supply chain, and find out the cooperative partners suitable for the strategical alliance of Rizhao port supply chain among different types of alliance. II. THE SWOT ANALYSIS OF RIZHAO PORT SUPPLY CHAIN The SWOT analysis, a commonly used method of strategic competition, is based on analyzing the internal conditions of enterprises itself, and finds out the strengths, weaknesses and core competence. -

Land-Use Efficiency in Shandong (China)
sustainability Article Land-Use Efficiency in Shandong (China): Empirical Analysis Based on a Super-SBM Model Yayuan Pang and Xinjun Wang * Department of Environmental Science and Engineering, Fudan University, Shanghai 200433, China; [email protected] * Correspondence: [email protected] Received: 20 November 2020; Accepted: 14 December 2020; Published: 18 December 2020 Abstract: A reasonable evaluation of land-use efficiency is an important issue in land use and development. By using a super-SBM model, the construction and cultivated land-use efficiency of 17 cities in Shandong from 2006 to 2018 were estimated and the spatial-temporal variation was analyzed. The results showed that: (1) The land use efficiency levels were quite different, and low-efficiency cities impacted the overall development process. (2) The efficiency values of construction land generally fluctuated and rose, meaning that room remains for future efficiency improvements. Cultivated land generally showed a high utilization efficiency, but it fluctuated and decreased. (3) The construction land-use efficiency was highest in the midland region, especially in Laiwu city, followed by the eastern region and Qingdao city, and the western region. The spatial variation in cultivated land presented a trend of “high in the middle, low in the periphery,” centered on Jinan and Yantai city. (4) Pure technical efficiency was the main restriction driving inefficient utilization in the western region, while scale efficiency played that role in the east. Based on the findings, policy suggestions were proposed to improve the land-use efficiency in Shandong and promote urban sustainable development. Keywords: land use; efficiency level; super-SBM model; Shandong Province; construction land; cultivated land 1. -

Spatial and Temporal Variation of VIIRS Derived AOT
Spatial and temporal variation of the Visible Infrared Imaging Radiometer Suite (VIIRS) derived aerosol optical thickness over East China Fei Menga, b, Changyong Caoc, Xi Shaoa aDepartment of Astronomy, University of Maryland, College Park, MD, USA bDepartment of Civil Engineering, Shandong Jianzhu University, Jinan, PR China cNOAA/ NESDIS/STAR, College Park, MD, USA Abstract Results The spatial and temporal variations in regional aerosol optical thickness (AOT) were investigated over Annual and seasonal AOT variations for different cities Shandong province of China based upon one year’s Visible Infrared Imaging Radiometer Suite (VIIRS) Averaged over the measurements in 2013, the minimum (Min.), maximum (Max.), mean, standard deviation data. The regional forest background annual mean AOT was 0.467 with a standard deviation of 0.339, (Std.) and variance of AOT in 17 cities (Fig.1) are presented in Table 1. which was much higher than the background continental AOT level of 0.10. Higher AOT values for the In order to better understand the AOT values in different cities, percent days with AOT≤0.5, 1.0≥AOT>0.5 and study region were mainly found in the spring and summer, especially from May to August, while the AOT>1.0 respectively in each cities were calculated (Fig.2). (a) (b) (c) (d) lowest mean aerosol values were seen in November and December. Urban areas all have obviously higher mean AOT values than the rural areas resulting from intense anthropogenic sources. Given that the forest background AOT represents the natural background level, anthropogenic emissions and C secondary aerosol generation contribute approximately 0.352 to the aerosol loading in this region. -
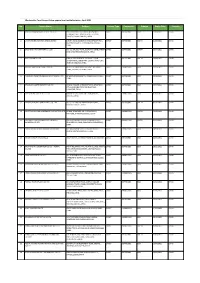
Apparel & Textile Factory List
Woolworths Food Group-Active apparel and textile factories- April 2021 No. Factory Name Address Factory Type Commodity Scheme Expiry Date Country 1 JIANGSU HONGMOFANG TEXTILE CO., LTD EAST CIFU ROAD, EAST RUISHENG AVENUE,, DIRECT SOFTGOODS BSCI 23/04/2021 CHINA ECONOMIC DEVELOPMENT ZONE, SHUYANG COUNTY,,SUQIAN,JIANGSU,,CHINA 2 LIANYUNGANG ZHAOWEN SHOES LIMITED 2 BEIHAI ROAD,ECONOMIC DEVELOPMENT AREA, DIRECT SOFTGOODS SMETA 11/05/2021 CHINA GUANNAN COUNTY,,LIANYUNGANG,JIANGSU,, CHINA 3 ZHUCHENG YIXIN GARMENT CO., LTD NO. 321 SHUNDU ROAD,,ECONOMIC DEVELOPMENT DIRECT SOFTGOODS SMETA 13/07/2021 CHINA ZONE,,ZHUCHENG,SHANDONG,,CHINA 4 HRX FASHION CO LTD 1000 METERS SOUTH OF GUOCANG TOWN DIRECT SOFTGOODS SMETA 25/08/2021 CHINA GOVERNMENT,,WENSHANG COUNTY, JINING CITY, JINING,SHANDONG,,CHINA 5 PUJIANG KINGSHOW CARPET CO LTD NO.75-1 ZHEN PU ROAD, PU JIANG, ZHE JIANG, DIRECT HARDGOODS BSCI 24/12/2021 CHINA CHINA,,PUJIANG,ZHEJIANG,,CHINA 6 YANGZHOU TENGYI SHOES MANUFACTURE CO., LTD. 13 WEST HONGCHENG RD,,,FANGXIANG,JIANGSU,, DIRECT SOFTGOODS BSCI 22/10/2021 CHINA CHINA 7 ZHAOYUAN CASTTE GARMENT CO LTD PANJIAJI VILLAGE, LINGLONG TOWN,,ZHAOYUAN DIRECT SOFTGOODS BSCI 06/07/2021 CHINA CITY, SHANDONG PROVINCE,ZHAOYUAN, SHANDONG,,CHINA 8 RUGAO HONGTAI TEXTILE CO LTD XINJIAN VILLAGE, JIANGAN TOWN,,RUGAO, DIRECT HARDGOODS BSCI 07/12/2021 CHINA JIANGSU,,CHINA 9 NANJING BIAOMEI HOMETEXTILES CO.,LTD NO.13 ERST WUCHU ROAD,HENGXI TOWN,,, DIRECT SOFTGOODS SMETA 04/11/2021 CHINA NANJING,JIANGSU,,CHINA 10 NANTONG YAOXING HOUSEWARE PRODUCTS CO.,LTD NO.999, TONGFUBEI RD., CHONGCHUAN, DIRECT HARDGOODS SMETA 24/09/2021 CHINA NANTONG,,NANTONG,JIANGSU,,CHINA 11 CHAOZHOU CHAOAN ZHENGYUN CERAMICS QIAO HU VILLAGE, CHAOAN, CHAOZHOU CITY, DIRECT HARDGOODS SMETA 19/05/2021 CHINA INDUSTRIAL CO LTD GUANGDONG, CHINA,,CHAOZHOU,GUANGDONG,, CHINA 12 YANTAI PACIFIC HOME FASHION FUSHAN MILL NO. -

Slip Op. 19-67 UNITED STATES COURT OF
Case 1:18-cv-00002-JCG Document 81 Filed 06/03/19 Page 1 of 30 Slip Op. 19-67 UNITED STATES COURT OF INTERNATIONAL TRADE LINYI CHENGEN IMPORT AND EXPORT CO., LTD., Plaintiff, and CELTIC CO., LTD. ET AL., Consolidated Plaintiffs, Before: Jennifer Choe-Groves, Judge v. Consol. Court No. 18-00002 UNITED STATES, Defendant, and COALITION FOR FAIR TRADE IN HARDWOOD PLYWOOD, Defendant-Intervenor. OPINION AND ORDER [Sustaining in part and remanding in part the U.S. Department of Commerce’s final determination in the antidumping duty investigation of certain hardwood plywood products from the People’s Republic of China.] Dated: June 3, 2019 Gregory S. Menegaz and Alexandra H. Salzman, deKieffer & Horgan, PLLC, of Washington, D.C., argued for Plaintiff Linyi Chengen Import and Export Co., Ltd., Consolidated Plaintiffs Celtic Co., Ltd., Jiaxing Gsun Import & Export Co., Ltd., Suqian Hopeway International Trade Co., Ltd., Anhui Hoda Wood Co., Ltd., Shanghai Futuwood Trading Co., Ltd., Linyi Evergreen Wood Co., Ltd., Xuzhou Jiangyang Wood Industries Co., Ltd., Xuzhou Timber International Trade Co. Ltd., Linyi Sanfortune Wood Co., Ltd., Linyi Mingzhu Wood Co., Ltd., Xuzhou Andefu Wood Co., Ltd., Suining Pengxiang Wood Co., Ltd., Xuzhou Shengping Import and Export Co., Ltd., Xuzhou Pinlin International Trade Co. Ltd., Linyi Glary Plywood Co., Ltd., Case 1:18-cv-00002-JCG Document 81 Filed 06/03/19 Page 2 of 30 Consol. Court No. 18-00002 Page 2 Linyi Linhai Wood Co., Ltd., Linyi Hengsheng Wood Industry Co., Ltd., Shandong Qishan International Trading -

Annual Report of Shandong Hi-Speed Co. Ltd. of 2020
Annual Report 2020 Company Code: 600350 Abbreviation of Company: Shangdong Hi-Speed Annual Report of Shandong Hi-Speed Co. Ltd. of 2020 1/265 Annual Report 2020 Notes I. The Board of Directors, Board of Supervisors, directors, supervisors and executives of the Company guarantee the truthfulness, accuracy and completeness without any false or misleading statements or material omissions herein, and shall bear joint and several legal liabilities. II. Absent directors Post of absent director Name of absent director Reason for absence Name of delegate Independent Director Fan Yuejin Because of work Wei Jian III. Shinewing Certified Public Accountants (Special Partnership) has issued an unqualified auditor's report for the Company. IV. Mr. Sai Zhiyi, the head of the Company, Mr. Lyu Sizhong, Chief Accountant who is in charge of accounting affairs, Mr. Zhou Liang, and Chen Fang (accountant in charge) from ShineWing declared to guarantee the truthfulness, accuracy and completeness of the annual report. V. With respect to the profit distribution plan or common reserves capitalizing plan during the reporting period reviewed by the Board of Directors After being audited by Shinewing Certified Public Accountants (Special Partnership), the net profit attributable to owners of the parent company in 2020 after consolidation is CNY 2,038,999,018.13, where: the net profit achieved by the parent company is CNY2,242,060,666.99. After withdrawing the statutory reserves of CNY224,206,066.70 at a ratio of 10% of the achieved net profit of the parent company, the retained earnings is 2,017,854,600.29 . The accumulated distributable profits of parent company in 2020 is CNY16,232,090,812.89.