Proteomics Analysis of the Proliferative Effect of Low-Dose Ouabain on Human Endothelial Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

KLF2 Induced
UvA-DARE (Digital Academic Repository) The transcription factor KLF2 in vascular biology Boon, R.A. Publication date 2008 Link to publication Citation for published version (APA): Boon, R. A. (2008). The transcription factor KLF2 in vascular biology. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:23 Sep 2021 Supplementary data: Genes induced by KLF2 Dekker et al. LocusLink Accession Gene Sequence Description Fold p-value ID number symbol change (FDR) 6654 AK022099 SOS1 cDNA FLJ12037 fis, clone HEMBB1001921. 100.00 5.9E-09 56999 AF086069 ADAMTS9 full length insert cDNA clone YZ35C05. 100.00 1.2E-09 6672 AF085934 SP100 full length insert cDNA clone YR57D07. 100.00 6.7E-13 9031 AF132602 BAZ1B Williams Syndrome critical region WS25 mRNA, partial sequence. -

Steroid-Dependent Regulation of the Oviduct: a Cross-Species Transcriptomal Analysis
University of Kentucky UKnowledge Theses and Dissertations--Animal and Food Sciences Animal and Food Sciences 2015 Steroid-dependent regulation of the oviduct: A cross-species transcriptomal analysis Katheryn L. Cerny University of Kentucky, [email protected] Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Cerny, Katheryn L., "Steroid-dependent regulation of the oviduct: A cross-species transcriptomal analysis" (2015). Theses and Dissertations--Animal and Food Sciences. 49. https://uknowledge.uky.edu/animalsci_etds/49 This Doctoral Dissertation is brought to you for free and open access by the Animal and Food Sciences at UKnowledge. It has been accepted for inclusion in Theses and Dissertations--Animal and Food Sciences by an authorized administrator of UKnowledge. For more information, please contact [email protected]. STUDENT AGREEMENT: I represent that my thesis or dissertation and abstract are my original work. Proper attribution has been given to all outside sources. I understand that I am solely responsible for obtaining any needed copyright permissions. I have obtained needed written permission statement(s) from the owner(s) of each third-party copyrighted matter to be included in my work, allowing electronic distribution (if such use is not permitted by the fair use doctrine) which will be submitted to UKnowledge as Additional File. I hereby grant to The University of Kentucky and its agents the irrevocable, non-exclusive, and royalty-free license to archive and make accessible my work in whole or in part in all forms of media, now or hereafter known. -

1 Metabolic Dysfunction Is Restricted to the Sciatic Nerve in Experimental
Page 1 of 255 Diabetes Metabolic dysfunction is restricted to the sciatic nerve in experimental diabetic neuropathy Oliver J. Freeman1,2, Richard D. Unwin2,3, Andrew W. Dowsey2,3, Paul Begley2,3, Sumia Ali1, Katherine A. Hollywood2,3, Nitin Rustogi2,3, Rasmus S. Petersen1, Warwick B. Dunn2,3†, Garth J.S. Cooper2,3,4,5* & Natalie J. Gardiner1* 1 Faculty of Life Sciences, University of Manchester, UK 2 Centre for Advanced Discovery and Experimental Therapeutics (CADET), Central Manchester University Hospitals NHS Foundation Trust, Manchester Academic Health Sciences Centre, Manchester, UK 3 Centre for Endocrinology and Diabetes, Institute of Human Development, Faculty of Medical and Human Sciences, University of Manchester, UK 4 School of Biological Sciences, University of Auckland, New Zealand 5 Department of Pharmacology, Medical Sciences Division, University of Oxford, UK † Present address: School of Biosciences, University of Birmingham, UK *Joint corresponding authors: Natalie J. Gardiner and Garth J.S. Cooper Email: [email protected]; [email protected] Address: University of Manchester, AV Hill Building, Oxford Road, Manchester, M13 9PT, United Kingdom Telephone: +44 161 275 5768; +44 161 701 0240 Word count: 4,490 Number of tables: 1, Number of figures: 6 Running title: Metabolic dysfunction in diabetic neuropathy 1 Diabetes Publish Ahead of Print, published online October 15, 2015 Diabetes Page 2 of 255 Abstract High glucose levels in the peripheral nervous system (PNS) have been implicated in the pathogenesis of diabetic neuropathy (DN). However our understanding of the molecular mechanisms which cause the marked distal pathology is incomplete. Here we performed a comprehensive, system-wide analysis of the PNS of a rodent model of DN. -

Anti-Inflammatory Role of Curcumin in LPS Treated A549 Cells at Global Proteome Level and on Mycobacterial Infection
Anti-inflammatory Role of Curcumin in LPS Treated A549 cells at Global Proteome level and on Mycobacterial infection. Suchita Singh1,+, Rakesh Arya2,3,+, Rhishikesh R Bargaje1, Mrinal Kumar Das2,4, Subia Akram2, Hossain Md. Faruquee2,5, Rajendra Kumar Behera3, Ranjan Kumar Nanda2,*, Anurag Agrawal1 1Center of Excellence for Translational Research in Asthma and Lung Disease, CSIR- Institute of Genomics and Integrative Biology, New Delhi, 110025, India. 2Translational Health Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, 110067, India. 3School of Life Sciences, Sambalpur University, Jyoti Vihar, Sambalpur, Orissa, 768019, India. 4Department of Respiratory Sciences, #211, Maurice Shock Building, University of Leicester, LE1 9HN 5Department of Biotechnology and Genetic Engineering, Islamic University, Kushtia- 7003, Bangladesh. +Contributed equally for this work. S-1 70 G1 S 60 G2/M 50 40 30 % of cells 20 10 0 CURI LPSI LPSCUR Figure S1: Effect of curcumin and/or LPS treatment on A549 cell viability A549 cells were treated with curcumin (10 µM) and/or LPS or 1 µg/ml for the indicated times and after fixation were stained with propidium iodide and Annexin V-FITC. The DNA contents were determined by flow cytometry to calculate percentage of cells present in each phase of the cell cycle (G1, S and G2/M) using Flowing analysis software. S-2 Figure S2: Total proteins identified in all the three experiments and their distribution betwee curcumin and/or LPS treated conditions. The proteins showing differential expressions (log2 fold change≥2) in these experiments were presented in the venn diagram and certain number of proteins are common in all three experiments. -

Visinin-Like Neuronal Calcium Sensor Proteins Regulate the Slow Calcium-Activated Afterhyperpolarizing Current in the Rat Cerebral Cortex
The Journal of Neuroscience, October 27, 2010 • 30(43):14361–14365 • 14361 Brief Communications Visinin-Like Neuronal Calcium Sensor Proteins Regulate the Slow Calcium-Activated Afterhyperpolarizing Current in the Rat Cerebral Cortex Claudio Villalobos and Rodrigo Andrade Department of Pharmacology, Wayne State University School of Medicine, Detroit Michigan 48230 Many neurons in the nervous systems express afterhyperpolarizations that are mediated by a slow calcium-activated potassium current. This current shapes neuronal firing and is inhibited by neuromodulators, suggesting an important role in the regulation of neuronal function. Surprisingly, very little is currently known about the molecular basis for this current or how it is gated by calcium. Recently, the neuronal calcium sensor protein hippocalcin was identified as a calcium sensor for the slow afterhyperpolarizing current in the hip- pocampus. However, while hippocalcin is very strongly expressed in the hippocampus, this protein shows a relatively restricted distri- bution in the brain. Furthermore, the genetic deletion of this protein only partly reduces the slow hyperpolarizing current in hippocampus. These considerations question whether hippocalcin can be the sole calcium sensor for the slow afterhyperpolarizing current. Here we use loss of function and overexpression strategies to show that hippocalcin functions as a calcium sensor for the slow afterhyperpolarizing current in the cerebral cortex, an area where hippocalcin is expressed at much lower levels than in hippocampus. In addition we show that neurocalcin ␦, but not VILIP-2, can also act as a calcium sensor for the slow afterhyperpolarizing current. Finally we show that hippocalcin and neurocalcin ␦ both increase the calcium sensitivity of the afterhyperpolarizing current but do not alter its ␣  sensitivity to inhibition by carbachol acting through the G q-11-PLC signaling cascade. -

Gene Expression Signatures and Biomarkers of Noninvasive And
Oncogene (2006) 25, 2328–2338 & 2006 Nature Publishing Group All rights reserved 0950-9232/06 $30.00 www.nature.com/onc ORIGINAL ARTICLE Gene expression signatures and biomarkers of noninvasive and invasive breast cancer cells: comprehensive profiles by representational difference analysis, microarrays and proteomics GM Nagaraja1, M Othman2, BP Fox1, R Alsaber1, CM Pellegrino3, Y Zeng2, R Khanna2, P Tamburini3, A Swaroop2 and RP Kandpal1 1Department of Biological Sciences, Fordham University, Bronx, NY, USA; 2Department of Ophthalmology and Visual Sciences, University of Michigan, Ann Arbor, MI, USA and 3Bayer Corporation, West Haven, CT, USA We have characterized comprehensive transcript and Keywords: representational difference analysis; micro- proteomic profiles of cell lines corresponding to normal arrays; proteomics; breast carcinoma; biomarkers; breast (MCF10A), noninvasive breast cancer (MCF7) and copper homeostasis invasive breast cancer (MDA-MB-231). The transcript profiles were first analysed by a modified protocol for representational difference analysis (RDA) of cDNAs between MCF7 and MDA-MB-231 cells. The majority of genes identified by RDA showed nearly complete con- Introduction cordance withmicroarray results, and also led to the identification of some differentially expressed genes such The transformation of a normal cell into a cancer cell as lysyl oxidase, copper transporter ATP7A, EphB6, has been correlated to altered expression of a variety of RUNX2 and a variant of RUNX2. The altered transcripts genes (Perou et al., 2000; Becker et al., 2005). The identified by microarray analysis were involved in cell–cell expression of some of these genes is a direct result of or cell–matrix interaction, Rho signaling, calcium home- sequence mutation, whereas other changes occur due to ostasis and copper-binding/sensitive activities. -

Proteomic Analysis of Exosome-Like Vesicles Derived from Breast Cancer Cells
ANTICANCER RESEARCH 32: 847-860 (2012) Proteomic Analysis of Exosome-like Vesicles Derived from Breast Cancer Cells GEMMA PALAZZOLO1, NADIA NINFA ALBANESE2,3, GIANLUCA DI CARA3, DANIEL GYGAX4, MARIA LETIZIA VITTORELLI3 and IDA PUCCI-MINAFRA3 1Institute for Biomedical Engineering, Laboratory of Biosensors and Bioelectronics, ETH Zurich, Switzerland; 2Department of Physics, University of Palermo, Palermo, Italy; 3Centro di Oncobiologia Sperimentale (C.OB.S.), Oncology Department La Maddalena, Palermo, Italy; 4Institute of Chemistry and Bioanalytics, University of Applied Sciences Northwestern Switzerland FHNW, Muttenz, Switzerland Abstract. Background/Aim: The phenomenon of membrane that vesicle production allows neoplastic cells to exert different vesicle-release by neoplastic cells is a growing field of interest effects, according to the possible acceptor targets. For instance, in cancer research, due to their potential role in carrying a vesicles could potentiate the malignant properties of adjacent large array of tumor antigens when secreted into the neoplastic cells or activate non-tumoral cells. Moreover, vesicles extracellular medium. In particular, experimental evidence show could convey signals to immune cells and surrounding stroma that at least some of the tumor markers detected in the blood cells. The present study may significantly contribute to the circulation of mammary carcinoma patients are carried by knowledge of the vesiculation phenomenon, which is a critical membrane-bound vesicles. Thus, biomarker research in breast device for trans cellular communication in cancer. cancer can gain great benefits from vesicle characterization. Materials and Methods: Conditioned medium was collected The phenomenon of membrane release in the extracellular from serum starved MDA-MB-231 sub-confluent cell cultures medium has long been known and was firstly described by and exosome-like vesicles (ELVs) were isolated by Paul H. -

Beta Cell Adaptation to Pregnancy Requires Prolactin Action on Both
www.nature.com/scientificreports OPEN Beta cell adaptation to pregnancy requires prolactin action on both beta and non‑beta cells Vipul Shrivastava1, Megan Lee1, Daniel Lee1, Marle Pretorius1, Bethany Radford1, Guneet Makkar1 & Carol Huang1,2,3* Pancreatic islets adapt to insulin resistance of pregnancy by up regulating β‑cell mass and increasing insulin secretion. Previously, using a transgenic mouse with global, heterozygous deletion of prolactin receptor (Prlr+/−), we found Prlr signaling is important for this adaptation. However, since Prlr is expressed in tissues outside of islets as well as within islets and prolactin signaling afects β‑cell development, to understand β‑cell‑specifc efect of prolactin signaling in pregnancy, we generated a transgenic mouse with an inducible conditional deletion of Prlr from β‑cells. Here, we found that β‑cell‑specifc Prlr reduction in adult mice led to elevated blood glucose, lowed β‑cell mass and blunted in vivo glucose‑stimulated insulin secretion during pregnancy. When we compared gene expression profle of islets from transgenic mice with global (Prlr+/−) versus β‑cell‑specifc Prlr reduction (βPrlR+/−), we found 95 diferentially expressed gene, most of them down regulated in the Prlr+/− mice in comparison to the βPrlR+/− mice, and many of these genes regulate apoptosis, synaptic vesicle function and neuronal development. Importantly, we found that islets from pregnant Prlr+/− mice are more vulnerable to glucolipotoxicity‑induced apoptosis than islets from pregnant βPrlR+/− mice. These observations suggest that down regulation of prolactin action during pregnancy in non‑β‑cells secondarily and negatively afect β‑cell gene expression, and increased β‑cell susceptibility to external insults. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -
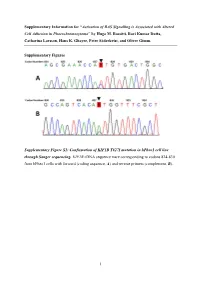
Activation of RAS Signalling Is Associated with Altered Cell Adhesion in Phaeochromocytoma” by Hugo M
Supplementary Information for “Activation of RAS Signalling is Associated with Altered Cell Adhesion in Phaeochromocytoma” by Hugo M. Rossitti, Ravi Kumar Dutta, Catharina Larsson, Hans K. Ghayee, Peter Söderkvist, and Oliver Gimm. Supplementary Figures Supplementary Figure S1: Confirmation of KIF1B T827I mutation in hPheo1 cell line through Sanger sequencing. KIF1B cDNA sequence trace corresponding to codons 824-830 from hPheo1 cells with forward (coding sequence, A) and reverse primers (complement, B). 1 Supplementary Figure S2: Confirmation of NRAS Q61K mutation in hPheo1 cell line through Sanger sequencing. NRAS cDNA sequence trace corresponding to codons 56-67 from hPheo1 cells with forward (coding sequence, A) and reverse primers (complement, B). 2 Supplementary Figure S3: CCND1 gene expression and hPheo1 proliferation. A: Expression of CCND1 mRNA assessed by RT-qPCR and presented as fold change (2-ΔΔCT, mean ± standard error of the mean). B: Cell counts at 1, 2, and 3 days after plating (corresponding to 72, 96 and 120 hours posttransfection, respectively) of control- or siNRAS#1-transfected hPheo1 cells expressed as fold change of the number of cells plated at day 0 (48 hours posttransfection; mean ± standard deviation). All results are from three independent siRNA experiments. 3 Supplementary Tables Supplementary Table S1: List of transcript cluster IDs significantly upregulated in hPheo1 by siNRAS treatment (comparison: siNRAS versus control-transfected hPheo1; ANOVA p < 0.05, FDR < 0.25, fold change < -1.5 or > 1.5). Transcript -

Gene Expression Profiling of Differentiated Thyroid Neoplasms: Diagnostic and Clinical Implications
6586 Vol. 10, 6586–6597, October 1, 2004 Clinical Cancer Research Gene Expression Profiling of Differentiated Thyroid Neoplasms: Diagnostic and Clinical Implications Sylvie Chevillard,1 Nicolas Ugolin,1 entiation, adhesion, immune response, and proliferation as- Philippe Vielh,2 Katherine Ory,1 Ce´line Levalois,1 sociated pathways. Quantitative real-time reverse transcrip- Danielle Elliott,4 Gary L. Clayman,3 and tion-PCR analysis of selected genes corroborated the 4 microarray expression results. Adel K. El-Naggar Conclusions: Our study show the following: (1) differ- 1 Laboratoire de Cance´rologie Expe´rimentale, Commissariat a´ ences in gene expression between tumor and nontumor bear- L’Energie Atomique, Direction des Sciences du Vivant, De´partement ing normal thyroid tissue can be identified, (2) a set of genes du Radiobiologie et Radiopathologie, Fontenay-aux-Roses Cedex France; 2De´partement de Pathologie, Institut Gustave Roussy, differentiate follicular neoplasm from follicular variant of Villejuif Cedex, France; Departments of 3Head and Neck Surgery and papillary carcinoma, (3) follicular adenoma and carcinoma 4Pathology, The University of Texas M.D. Anderson Cancer Center, share many of the differentiated genes, and (4) gene expres- Houston, Texas sion differences identify conventional papillary carcinoma from the follicular variant. ABSTRACT Purpose: The purpose of this research was to identify INTRODUCTION novel genes that can be targeted as diagnostic and clinical Differentiated thyroid epithelial tumors represent a spec- markers of differentiated thyroid tumors. trum of morphologically and biologically diverse lesions. The Experimental Design: Gene expression analysis using follicular-derived neoplasms (adenoma, carcinoma, and the fol- microarray platform was performed on 6 pathologically licular variant of papillary carcinoma) manifest overlapping normal thyroid samples and 12 primary follicular and pap- cytomorphologic features and not infrequently pose diagnostic illary thyroid neoplasms. -

Probe Set Name Symbol 1598 G at Growth Arres
Supplementary Table 2. List of stroma related genes (i.e. probe sets overexpressed in core relative to FNA biopsies of the same cancer) Probe set Name Symbol 1598_g_at growth arrest-specific 6 GAS6 200048_s_at jumping translocation breakpoint JTB 200054_at zinc finger protein 259 ZNF259 200055_at TAF10 RNA polymerase II, TATA box binding protein (TBP)-associatedTAF10 factor, 30kDa 200059_s_at ras homolog gene family, member A RHOA 200060_s_at RNA binding protein S1, serine-rich domain RNPS1 200070_at chromosome 2 open reading frame 24 C2orf24 200613_at adaptor-related protein complex 2, mu 1 subunit AP2M1 200663_at CD63 molecule CD63 200665_s_at secreted protein, acidic, cysteine-rich (osteonectin) SPARC 200671_s_at spectrin, beta, non-erythrocytic 1 SPTBN1 200696_s_at gelsolin (amyloidosis, Finnish type) GSN 200704_at lipopolysaccharide-induced TNF factor LITAF 200738_s_at phosphoglycerate kinase 1 PGK1 200760_s_at ADP-ribosylation-like factor 6 interacting protein 5 ARL6IP5 200762_at dihydropyrimidinase-like 2 DPYSL2 200770_s_at laminin, gamma 1 (formerly LAMB2) LAMC1 200771_at laminin, gamma 1 (formerly LAMB2) LAMC1 200772_x_at prothymosin, alpha PTMA 200778_s_at septin 2 2-Sep 200782_at annexin A5 ANXA5 200784_s_at low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor)LRP1 200785_s_at low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor)LRP1 200795_at SPARC-like 1 (hevin) SPARCL1 200799_at heat shock 70kDa protein 1A HSPA1A 200807_s_at heat shock 60kDa protein 1 (chaperonin) HSPD1 200811_at