Evaluation of the Disconnect Between Hepatocyte and Microsome Intrinsic Clearance and in Vitro in Vivo Extrapolation Performance
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(CD-P-PH/PHO) Report Classification/Justifica
COMMITTEE OF EXPERTS ON THE CLASSIFICATION OF MEDICINES AS REGARDS THEIR SUPPLY (CD-P-PH/PHO) Report classification/justification of medicines belonging to the ATC group R01 (Nasal preparations) Table of Contents Page INTRODUCTION 5 DISCLAIMER 7 GLOSSARY OF TERMS USED IN THIS DOCUMENT 8 ACTIVE SUBSTANCES Cyclopentamine (ATC: R01AA02) 10 Ephedrine (ATC: R01AA03) 11 Phenylephrine (ATC: R01AA04) 14 Oxymetazoline (ATC: R01AA05) 16 Tetryzoline (ATC: R01AA06) 19 Xylometazoline (ATC: R01AA07) 20 Naphazoline (ATC: R01AA08) 23 Tramazoline (ATC: R01AA09) 26 Metizoline (ATC: R01AA10) 29 Tuaminoheptane (ATC: R01AA11) 30 Fenoxazoline (ATC: R01AA12) 31 Tymazoline (ATC: R01AA13) 32 Epinephrine (ATC: R01AA14) 33 Indanazoline (ATC: R01AA15) 34 Phenylephrine (ATC: R01AB01) 35 Naphazoline (ATC: R01AB02) 37 Tetryzoline (ATC: R01AB03) 39 Ephedrine (ATC: R01AB05) 40 Xylometazoline (ATC: R01AB06) 41 Oxymetazoline (ATC: R01AB07) 45 Tuaminoheptane (ATC: R01AB08) 46 Cromoglicic Acid (ATC: R01AC01) 49 2 Levocabastine (ATC: R01AC02) 51 Azelastine (ATC: R01AC03) 53 Antazoline (ATC: R01AC04) 56 Spaglumic Acid (ATC: R01AC05) 57 Thonzylamine (ATC: R01AC06) 58 Nedocromil (ATC: R01AC07) 59 Olopatadine (ATC: R01AC08) 60 Cromoglicic Acid, Combinations (ATC: R01AC51) 61 Beclometasone (ATC: R01AD01) 62 Prednisolone (ATC: R01AD02) 66 Dexamethasone (ATC: R01AD03) 67 Flunisolide (ATC: R01AD04) 68 Budesonide (ATC: R01AD05) 69 Betamethasone (ATC: R01AD06) 72 Tixocortol (ATC: R01AD07) 73 Fluticasone (ATC: R01AD08) 74 Mometasone (ATC: R01AD09) 78 Triamcinolone (ATC: R01AD11) 82 -
Prohibited Substances List
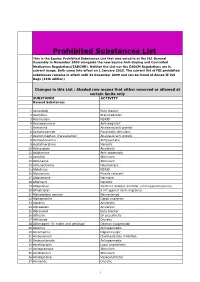
Prohibited Substances List This is the Equine Prohibited Substances List that was voted in at the FEI General Assembly in November 2009 alongside the new Equine Anti-Doping and Controlled Medication Regulations(EADCMR). Neither the List nor the EADCM Regulations are in current usage. Both come into effect on 1 January 2010. The current list of FEI prohibited substances remains in effect until 31 December 2009 and can be found at Annex II Vet Regs (11th edition) Changes in this List : Shaded row means that either removed or allowed at certain limits only SUBSTANCE ACTIVITY Banned Substances 1 Acebutolol Beta blocker 2 Acefylline Bronchodilator 3 Acemetacin NSAID 4 Acenocoumarol Anticoagulant 5 Acetanilid Analgesic/anti-pyretic 6 Acetohexamide Pancreatic stimulant 7 Acetominophen (Paracetamol) Analgesic/anti-pyretic 8 Acetophenazine Antipsychotic 9 Acetylmorphine Narcotic 10 Adinazolam Anxiolytic 11 Adiphenine Anti-spasmodic 12 Adrafinil Stimulant 13 Adrenaline Stimulant 14 Adrenochrome Haemostatic 15 Alclofenac NSAID 16 Alcuronium Muscle relaxant 17 Aldosterone Hormone 18 Alfentanil Narcotic 19 Allopurinol Xanthine oxidase inhibitor (anti-hyperuricaemia) 20 Almotriptan 5 HT agonist (anti-migraine) 21 Alphadolone acetate Neurosteriod 22 Alphaprodine Opiod analgesic 23 Alpidem Anxiolytic 24 Alprazolam Anxiolytic 25 Alprenolol Beta blocker 26 Althesin IV anaesthetic 27 Althiazide Diuretic 28 Altrenogest (in males and gelidngs) Oestrus suppression 29 Alverine Antispasmodic 30 Amantadine Dopaminergic 31 Ambenonium Cholinesterase inhibition 32 Ambucetamide Antispasmodic 33 Amethocaine Local anaesthetic 34 Amfepramone Stimulant 35 Amfetaminil Stimulant 36 Amidephrine Vasoconstrictor 37 Amiloride Diuretic 1 Prohibited Substances List This is the Equine Prohibited Substances List that was voted in at the FEI General Assembly in November 2009 alongside the new Equine Anti-Doping and Controlled Medication Regulations(EADCMR). -

Potential Drug Interactions and Potentially Inappropriate Medications in Daily Radiooncology Practice : a Risk Assessment
From the Department of Radiotherapy and Radiation Oncology of Marienhospital Herne – University Hospital – of the Ruhr-Universität Bochum Director: Prof. Dr. med. I. A. Adamietz Potential drug interactions and potentially inappropriate medications in daily radiooncology practice – a risk assessment Inaugural-Dissertation for the Attainment of a Doctor´s Degree in Medicine at the high Medical Faculty of the Ruhr-Universität Bochum Presented by Nina Sibylle Lamberty from Düsseldorf 2013 Dean of Faculty: Prof. Dr. med Klaus Überla Referee: Prof. Dr. med. Irenäus A. Adamietz Co-Referee: PD Dr. med Oliver Lindner Date of oral exam: 03.02.2015 Abstract Lamberty Nina Potential drug interactions and potentially inappropriate medications in daily radiooncology practice – a risk assess- ment Background: Radiation treatment of malignant disease includes patients with various extents of tumor and different comorbidities. Due to bad general condition approximately 20 % of irradiated subjects are in-patients. Beyond elimi- nation of tumor and treatment of its side effects drug interactions and potentially inappropriate medications constitute a major challenge in these radiooncologic patients. The layering of polypharmacy with age and radiation related physiological and functional changes and comorbidities might increase the risk for potential drug drug interactions (pDDs). This study attempted to quantify the frequency of pDDIs and potential inappropriate medications (PIMs) not related to i.v. chemotherapy in patients undergoing radiation treatment aiming the assessment of risk by medication. Methods: Medication profiles were analyzed by reviewing discharge letters of 120 cancer patients who had been admitted to the Marienhospital, Herne between November 2010 and November 2011. An improvement of the Karnofsky index during hospital treatment had been registered. -

Medicines Classification Committee
Medicines Classification Committee Meeting date 1 May 2017 58th Meeting Title Reclassification of Sedating Antihistamines Medsafe Pharmacovigilance Submitted by Paper type For decision Team Proposal for The Medicines Adverse Reactions Committee (MARC) recommended that the reclassification to committee consider reclassifying sedating antihistamines to prescription prescription medicines when used in children under 6 years of age for the treatment of medicine for some nausea and vomiting and travel sickness [exact wording to be determined by indications the committee]. Reason for The purpose of this document is to provide the committee with an overview submission of the information provided to the MARC about safety concerns associated with sedating antihistamines and reasons for recommendations. Associated March 2013 Children and Sedating Antihistamines Prescriber Update articles February 2010 Cough and cold medicines clarification – antihistamines Medsafe website Safety information: Use of cough and cold medicines in children – new advice Medicines for Alimemazine Diphenhydramine consideration Brompheniramine Doxylamine Chlorpheniramine Meclozine Cyclizine Promethazine Dexchlorpheniramine New Zealand Some oral sedating antihistamines available without exposure to a prescription (pharmacist-only and pharmacy only), sedating therefore usage data is not easily available. antihistamines Table of Contents 1.0 PURPOSE ...................................................................................................................................... -

Marrakesh Agreement Establishing the World Trade Organization
No. 31874 Multilateral Marrakesh Agreement establishing the World Trade Organ ization (with final act, annexes and protocol). Concluded at Marrakesh on 15 April 1994 Authentic texts: English, French and Spanish. Registered by the Director-General of the World Trade Organization, acting on behalf of the Parties, on 1 June 1995. Multilat ral Accord de Marrakech instituant l©Organisation mondiale du commerce (avec acte final, annexes et protocole). Conclu Marrakech le 15 avril 1994 Textes authentiques : anglais, français et espagnol. Enregistré par le Directeur général de l'Organisation mondiale du com merce, agissant au nom des Parties, le 1er juin 1995. Vol. 1867, 1-31874 4_________United Nations — Treaty Series • Nations Unies — Recueil des Traités 1995 Table of contents Table des matières Indice [Volume 1867] FINAL ACT EMBODYING THE RESULTS OF THE URUGUAY ROUND OF MULTILATERAL TRADE NEGOTIATIONS ACTE FINAL REPRENANT LES RESULTATS DES NEGOCIATIONS COMMERCIALES MULTILATERALES DU CYCLE D©URUGUAY ACTA FINAL EN QUE SE INCORPOR N LOS RESULTADOS DE LA RONDA URUGUAY DE NEGOCIACIONES COMERCIALES MULTILATERALES SIGNATURES - SIGNATURES - FIRMAS MINISTERIAL DECISIONS, DECLARATIONS AND UNDERSTANDING DECISIONS, DECLARATIONS ET MEMORANDUM D©ACCORD MINISTERIELS DECISIONES, DECLARACIONES Y ENTEND MIENTO MINISTERIALES MARRAKESH AGREEMENT ESTABLISHING THE WORLD TRADE ORGANIZATION ACCORD DE MARRAKECH INSTITUANT L©ORGANISATION MONDIALE DU COMMERCE ACUERDO DE MARRAKECH POR EL QUE SE ESTABLECE LA ORGANIZACI N MUND1AL DEL COMERCIO ANNEX 1 ANNEXE 1 ANEXO 1 ANNEX -

Vr Meds Ex01 3B 0825S Coding Manual Supplement Page 1
vr_meds_ex01_3b_0825s Coding Manual Supplement MEDNAME OTHER_CODE ATC_CODE SYSTEM THER_GP PHRM_GP CHEM_GP SODIUM FLUORIDE A12CD01 A01AA01 A A01 A01A A01AA SODIUM MONOFLUOROPHOSPHATE A12CD02 A01AA02 A A01 A01A A01AA HYDROGEN PEROXIDE D08AX01 A01AB02 A A01 A01A A01AB HYDROGEN PEROXIDE S02AA06 A01AB02 A A01 A01A A01AB CHLORHEXIDINE B05CA02 A01AB03 A A01 A01A A01AB CHLORHEXIDINE D08AC02 A01AB03 A A01 A01A A01AB CHLORHEXIDINE D09AA12 A01AB03 A A01 A01A A01AB CHLORHEXIDINE R02AA05 A01AB03 A A01 A01A A01AB CHLORHEXIDINE S01AX09 A01AB03 A A01 A01A A01AB CHLORHEXIDINE S02AA09 A01AB03 A A01 A01A A01AB CHLORHEXIDINE S03AA04 A01AB03 A A01 A01A A01AB AMPHOTERICIN B A07AA07 A01AB04 A A01 A01A A01AB AMPHOTERICIN B G01AA03 A01AB04 A A01 A01A A01AB AMPHOTERICIN B J02AA01 A01AB04 A A01 A01A A01AB POLYNOXYLIN D01AE05 A01AB05 A A01 A01A A01AB OXYQUINOLINE D08AH03 A01AB07 A A01 A01A A01AB OXYQUINOLINE G01AC30 A01AB07 A A01 A01A A01AB OXYQUINOLINE R02AA14 A01AB07 A A01 A01A A01AB NEOMYCIN A07AA01 A01AB08 A A01 A01A A01AB NEOMYCIN B05CA09 A01AB08 A A01 A01A A01AB NEOMYCIN D06AX04 A01AB08 A A01 A01A A01AB NEOMYCIN J01GB05 A01AB08 A A01 A01A A01AB NEOMYCIN R02AB01 A01AB08 A A01 A01A A01AB NEOMYCIN S01AA03 A01AB08 A A01 A01A A01AB NEOMYCIN S02AA07 A01AB08 A A01 A01A A01AB NEOMYCIN S03AA01 A01AB08 A A01 A01A A01AB MICONAZOLE A07AC01 A01AB09 A A01 A01A A01AB MICONAZOLE D01AC02 A01AB09 A A01 A01A A01AB MICONAZOLE G01AF04 A01AB09 A A01 A01A A01AB MICONAZOLE J02AB01 A01AB09 A A01 A01A A01AB MICONAZOLE S02AA13 A01AB09 A A01 A01A A01AB NATAMYCIN A07AA03 A01AB10 A A01 -

Respiratory Physiology Department
Patient Information Respiratory Physiology Department Skin Prick Testing What is a skin prick test? Your doctor has requested that you have a skin prick test performed to establish if you are allergic to any common allergens. This test is performed due to symptoms you have described to your doctor such as hay fever type symptoms, sneezing, red itchy eyes, itching or a wheeze. If the skin is exposed to an allergen, most patients who are sensitive to the particular allergen will usually produce a typical redness (erythema) and a raised weal (bump) within a few minutes of contact. When a reaction is produced it may help to assess the degree of sensitivity which is dependent of the size of the weal and the extent of the erythema. Pre-test instructions Before your appointment please stop taking the following medication for the stated time given prior to testing: Antihistamines Before your appointment stop for: Dimetindene Maleato 3 Days Fexofenadine 3 Days Clemastine 4 Days Cetirizine 3-10 Days Loratadine 3-10 Days Oxatomide 3-10 Days Chlorfeniramine 2-6 Days Patient Information Hydroxizine 4-6 Days Ketotifene 2 week Astemizole 2-4 Weeks Prednisolone >10mg 1 Week If you have had an allergic reaction within the two weeks prior to your appointment please contact us as the test results may not be accurate. Also, if you have an infection at the time of the test your system may be more responsive than usual; therefore the results may not give an accurate result. Performing the test When performing the test we use the following listed allergens: -

Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DIX to the HTSUS—Continued
20558 Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DEPARMENT OF THE TREASURY Services, U.S. Customs Service, 1301 TABLE 1.ÐPHARMACEUTICAL APPEN- Constitution Avenue NW, Washington, DIX TO THE HTSUSÐContinued Customs Service D.C. 20229 at (202) 927±1060. CAS No. Pharmaceutical [T.D. 95±33] Dated: April 14, 1995. 52±78±8 ..................... NORETHANDROLONE. A. W. Tennant, 52±86±8 ..................... HALOPERIDOL. Pharmaceutical Tables 1 and 3 of the Director, Office of Laboratories and Scientific 52±88±0 ..................... ATROPINE METHONITRATE. HTSUS 52±90±4 ..................... CYSTEINE. Services. 53±03±2 ..................... PREDNISONE. 53±06±5 ..................... CORTISONE. AGENCY: Customs Service, Department TABLE 1.ÐPHARMACEUTICAL 53±10±1 ..................... HYDROXYDIONE SODIUM SUCCI- of the Treasury. NATE. APPENDIX TO THE HTSUS 53±16±7 ..................... ESTRONE. ACTION: Listing of the products found in 53±18±9 ..................... BIETASERPINE. Table 1 and Table 3 of the CAS No. Pharmaceutical 53±19±0 ..................... MITOTANE. 53±31±6 ..................... MEDIBAZINE. Pharmaceutical Appendix to the N/A ............................. ACTAGARDIN. 53±33±8 ..................... PARAMETHASONE. Harmonized Tariff Schedule of the N/A ............................. ARDACIN. 53±34±9 ..................... FLUPREDNISOLONE. N/A ............................. BICIROMAB. 53±39±4 ..................... OXANDROLONE. United States of America in Chemical N/A ............................. CELUCLORAL. 53±43±0 -

Oral Disintegrating Composition of Anti-Histamine Agents
(19) TZZ 58¥6A_T (11) EP 2 589 376 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.05.2013 Bulletin 2013/19 A61K 9/00 (2006.01) A61K 9/20 (2006.01) A61K 31/445 (2006.01) (21) Application number: 12151435.0 (22) Date of filing: 17.01.2012 (84) Designated Contracting States: (72) Inventors: AL AT BE BG CH CY CZ DE DK EE ES FI FR GB •Raju,NS V GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO 560035 BANGALORE (IN) PL PT RO RS SE SI SK SM TR • Raghupathi, Kandarapu Designated Extension States: HYDERABAD (IN) BA ME • Rao Maram, Sambasiva 522601 Narasaraopet (IN) (30) Priority: 01.11.2011 IN CH37342011 (74) Representative: Regimbeau (71) Applicant: Inopharm Limited 20, rue de Chazelles 1304 Nicosia (CY) 75847 Paris Cedex 17 (FR) (54) Oral disintegrating composition of anti-histamine agents (57) The present invention is directed to an orally dis- more than 20, and one or more pharmaceutically accept- integrating composition of anti-histamine drug, wherein able excipients. the composition comprises anti-histamine drug, sur- factant of Hydrophilic Lipophilic Balance (HLB) value EP 2 589 376 A1 Printed by Jouve, 75001 PARIS (FR) EP 2 589 376 A1 Description [0001] The presentinvention relates tooral disintegrating composition ofanti-histamine agents. Particularly, the present invention relates to oral disintegrating composition of ebastine. More particularly, the present invention relates to 5 non-lyophilized composition of ebastine and its process for preparing the same. [0002] Ebastine is a long-acting and selective H1-histamine receptor antagonist. -

PHARMACEUTICAL APPENDIX to the HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2008) (Rev
Harmonized Tariff Schedule of the United States (2008) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2008) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. ABACAVIR 136470-78-5 ACIDUM GADOCOLETICUM 280776-87-6 ABAFUNGIN 129639-79-8 ACIDUM LIDADRONICUM 63132-38-7 ABAMECTIN 65195-55-3 ACIDUM SALCAPROZICUM 183990-46-7 ABANOQUIL 90402-40-7 ACIDUM SALCLOBUZICUM 387825-03-8 ABAPERIDONUM 183849-43-6 ACIFRAN 72420-38-3 ABARELIX 183552-38-7 ACIPIMOX 51037-30-0 ABATACEPTUM 332348-12-6 ACITAZANOLAST 114607-46-4 ABCIXIMAB 143653-53-6 ACITEMATE 101197-99-3 ABECARNIL 111841-85-1 ACITRETIN 55079-83-9 ABETIMUSUM 167362-48-3 ACIVICIN 42228-92-2 ABIRATERONE 154229-19-3 ACLANTATE 39633-62-0 ABITESARTAN 137882-98-5 ACLARUBICIN 57576-44-0 ABLUKAST 96566-25-5 ACLATONIUM NAPADISILATE 55077-30-0 ABRINEURINUM 178535-93-8 ACODAZOLE 79152-85-5 ABUNIDAZOLE 91017-58-2 ACOLBIFENUM 182167-02-8 ACADESINE 2627-69-2 ACONIAZIDE 13410-86-1 ACAMPROSATE -

2 12/ 35 74Al
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date 22 March 2012 (22.03.2012) 2 12/ 35 74 Al (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every A61K 9/16 (2006.01) A61K 9/51 (2006.01) kind of national protection available): AE, AG, AL, AM, A61K 9/14 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, (21) International Application Number: DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, PCT/EP201 1/065959 HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, (22) International Filing Date: KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, 14 September 201 1 (14.09.201 1) ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, (25) Filing Language: English RW, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, (26) Publication Language: English TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: 61/382,653 14 September 2010 (14.09.2010) US (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (71) Applicant (for all designated States except US): GM, KE, LR, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, NANOLOGICA AB [SE/SE]; P.O Box 8182, S-104 20 ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, Stockholm (SE). -

An Anticholinergic Burden Score for German Prescribers: Score Development Esther Katharina Kiesel1* , Yvonne Marina Hopf1 and Michael Drey2
Kiesel et al. BMC Geriatrics (2018) 18:239 https://doi.org/10.1186/s12877-018-0929-6 RESEARCH ARTICLE Open Access An anticholinergic burden score for German prescribers: score development Esther Katharina Kiesel1* , Yvonne Marina Hopf1 and Michael Drey2 Abstract Background: Anticholinergic drugs put elderly patients at a higher risk for falls, cognitive decline, and delirium as well as peripheral adverse reactions like dry mouth or constipation. Prescribers are often unaware of the drug- based anticholinergic burden (ACB) of their patients. This study aimed to develop an anticholinergic burden score for drugs licensed in Germany to be used by clinicians at prescribing level. Methods: A systematic literature search in pubmed assessed previously published ACB tools. Quantitative grading scores were extracted, reduced to drugs available in Germany, and reevaluated by expert discussion. Drugs were scored as having no, weak, moderate, or strong anticholinergic effects. Further drugs were identified in clinical routine and included as well. Results: The literature search identified 692 different drugs, with 548 drugs available in Germany. After exclusion of drugs due to no systemic effect or scoring of drug combinations (n = 67) and evaluation of 26 additional identified drugs in clinical routine, 504 drugs were scored. Of those, 356 drugs were categorised as having no, 104 drugs were scored as weak, 18 as moderate and 29 as having strong anticholinergic effects. Conclusions: The newly created ACB score for drugs authorized in Germany can be used in daily clinical practice to reduce potentially inappropriate medications for elderly patients. Further clinical studies investigating its effect on reducing anticholinergic side effects are necessary for validation.