Peroxisome Turnover and Diurnal Modulation of Antioxidant
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PEX5 Regulates Autophagy Via the Mtorc1-TFEB Axis During Starvation
Eun et al. Experimental & Molecular Medicine (2018) 50:4 DOI 10.1038/s12276-017-0007-8 Experimental & Molecular Medicine ARTICLE Open Access PEX5 regulates autophagy via the mTORC1-TFEB axis during starvation So Young Eun1,JoonNoLee2,In-KooNam2, Zhi-qiang Liu1,Hong-SeobSo 1, Seong-Kyu Choe1 and RaeKil Park2 Abstract Defects in the PEX5 gene impair the import of peroxisomal matrix proteins, leading to nonfunctional peroxisomes and other associated pathological defects such as Zellweger syndrome. Although PEX5 regulates autophagy process in a stress condition, the mechanisms controlling autophagy by PEX5 under nutrient deprivation are largely unknown. Herein, we show a novel function of PEX5 in the regulation of autophagy via Transcription Factor EB (TFEB). Under serum-starved conditions, when PEX5 is depleted, the mammalian target of rapamycin (mTORC1) inhibitor TSC2 is downregulated, which results in increased phosphorylation of the mTORC1 substrates, including 70S6K, S6K, and 4E- BP-1. mTORC1 activation further suppresses the nuclear localization of TFEB, as indicated by decreased mRNA levels of TFEB, LIPA, and LAMP1. Interestingly, peroxisomal mRNA and protein levels are also reduced by TFEB inactivation, indicating that TFEB might control peroxisome biogenesis at a transcriptional level. Conversely, pharmacological inhibition of mTOR resulting from PEX5 depletion during nutrient starvation activates TFEB by promoting nuclear localization of the protein. In addition, mTORC1 inhibition recovers the damaged-peroxisome biogenesis. These data suggest that PEX5 may be a critical regulator of lysosomal gene expression and autophagy through the mTOR-TFEB- autophagy axis under nutrient deprivation. 1234567890():,; 1234567890():,; Introduction Mitochondrial antiviral-signaling protein (MAVS) func- Peroxisome is an essential cellular organelle for per- tions as an antiviral signaling platform to induce the forming various metabolic activities, including oxidation interferon-independent signaling pathways4. -

Peroxisomal Disorders and Their Mouse Models Point to Essential Roles of Peroxisomes for Retinal Integrity
International Journal of Molecular Sciences Review Peroxisomal Disorders and Their Mouse Models Point to Essential Roles of Peroxisomes for Retinal Integrity Yannick Das, Daniëlle Swinkels and Myriam Baes * Lab of Cell Metabolism, Department of Pharmaceutical and Pharmacological Sciences, KU Leuven, 3000 Leuven, Belgium; [email protected] (Y.D.); [email protected] (D.S.) * Correspondence: [email protected] Abstract: Peroxisomes are multifunctional organelles, well known for their role in cellular lipid homeostasis. Their importance is highlighted by the life-threatening diseases caused by peroxisomal dysfunction. Importantly, most patients suffering from peroxisomal biogenesis disorders, even those with a milder disease course, present with a number of ocular symptoms, including retinopathy. Patients with a selective defect in either peroxisomal α- or β-oxidation or ether lipid synthesis also suffer from vision problems. In this review, we thoroughly discuss the ophthalmological pathology in peroxisomal disorder patients and, where possible, the corresponding animal models, with a special emphasis on the retina. In addition, we attempt to link the observed retinal phenotype to the underlying biochemical alterations. It appears that the retinal pathology is highly variable and the lack of histopathological descriptions in patients hampers the translation of the findings in the mouse models. Furthermore, it becomes clear that there are still large gaps in the current knowledge on the contribution of the different metabolic disturbances to the retinopathy, but branched chain fatty acid accumulation and impaired retinal PUFA homeostasis are likely important factors. Citation: Das, Y.; Swinkels, D.; Baes, Keywords: peroxisome; Zellweger; metabolism; fatty acid; retina M. Peroxisomal Disorders and Their Mouse Models Point to Essential Roles of Peroxisomes for Retinal Integrity. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Lipid Deposition and Metabolism in Local and Modern Pig Breeds: a Review
animals Review Lipid Deposition and Metabolism in Local and Modern Pig Breeds: A Review Klavdija Poklukar 1 , Marjeta Candek-Potokarˇ 1,2 , Nina Batorek Lukaˇc 1 , Urška Tomažin 1 and Martin Škrlep 1,* 1 Agricultural Institute of Slovenia, Ljubljana SI-1000, Slovenia; [email protected] (K.P.); [email protected] (M.C.-P.);ˇ [email protected] (N.B.L.); [email protected] (U.T.) 2 University of Maribor, Faculty of Agriculture and Life Sciences, HoˇceSI-2311, Slovenia * Correspondence: [email protected]; Tel.: +386-(0)1-280-52-34 Received: 17 February 2020; Accepted: 29 February 2020; Published: 3 March 2020 Simple Summary: Intensive selective breeding and genetic improvement of relatively few pig breeds led to the abandonment of many low productive local pig breeds. However, local pig breeds are more highly adapted to their specific environmental conditions and feeding resources, and therefore present a valuable genetic resource. They are able to deposit more fat and have a distinct lipogenic capacity, along with a better fatty acid composition than modern breeds. Physiological, biochemical and genetic mechanisms responsible for the differences between fatty and lean breeds are still not fully clarified. The present paper highlights important associations to better understand the underlying mechanisms of lipid deposition in subcutaneous and intramuscular fat between fatty and lean breeds. Abstract: Modern pig breeds, which have been genetically improved to achieve fast growth and a lean meat deposition, differ from local pig breeds with respect to fat deposition, fat specific metabolic characteristics and various other properties. The present review aimed to elucidate the mechanisms underlying the differences between fatty local and modern lean pig breeds in adipose tissue deposition and lipid metabolism, taking into consideration morphological, cellular, biochemical, transcriptomic and proteomic perspectives. -

Application of Microrna Database Mining in Biomarker Discovery and Identification of Therapeutic Targets for Complex Disease
Article Application of microRNA Database Mining in Biomarker Discovery and Identification of Therapeutic Targets for Complex Disease Jennifer L. Major, Rushita A. Bagchi * and Julie Pires da Silva * Department of Medicine, Division of Cardiology, University of Colorado Anschutz Medical Campus, Aurora, CO 80045, USA; [email protected] * Correspondence: [email protected] (R.A.B.); [email protected] (J.P.d.S.) Supplementary Tables Methods Protoc. 2021, 4, 5. https://doi.org/10.3390/mps4010005 www.mdpi.com/journal/mps Methods Protoc. 2021, 4, 5. https://doi.org/10.3390/mps4010005 2 of 25 Table 1. List of all hsa-miRs identified by Human microRNA Disease Database (HMDD; v3.2) analysis. hsa-miRs were identified using the term “genetics” and “circulating” as input in HMDD. Targets CAD hsa-miR-1 Targets IR injury hsa-miR-423 Targets Obesity hsa-miR-499 hsa-miR-146a Circulating Obesity Genetics CAD hsa-miR-423 hsa-miR-146a Circulating CAD hsa-miR-149 hsa-miR-499 Circulating IR Injury hsa-miR-146a Circulating Obesity hsa-miR-122 Genetics Stroke Circulating CAD hsa-miR-122 Circulating Stroke hsa-miR-122 Genetics Obesity Circulating Stroke hsa-miR-26b hsa-miR-17 hsa-miR-223 Targets CAD hsa-miR-340 hsa-miR-34a hsa-miR-92a hsa-miR-126 Circulating Obesity Targets IR injury hsa-miR-21 hsa-miR-423 hsa-miR-126 hsa-miR-143 Targets Obesity hsa-miR-21 hsa-miR-223 hsa-miR-34a hsa-miR-17 Targets CAD hsa-miR-223 hsa-miR-92a hsa-miR-126 Targets IR injury hsa-miR-155 hsa-miR-21 Circulating CAD hsa-miR-126 hsa-miR-145 hsa-miR-21 Targets Obesity hsa-mir-223 hsa-mir-499 hsa-mir-574 Targets IR injury hsa-mir-21 Circulating IR injury Targets Obesity hsa-mir-21 Targets CAD hsa-mir-22 hsa-mir-133a Targets IR injury hsa-mir-155 hsa-mir-21 Circulating Stroke hsa-mir-145 hsa-mir-146b Targets Obesity hsa-mir-21 hsa-mir-29b Methods Protoc. -

Rapid Whole Genome Sequencing Identifies a Homozygous Novel Variant, His540arg, in HSD17B4 Resulting in D-Bifunctional Protein Deficiency Disorder Diagnosis
Downloaded from molecularcasestudies.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press CSH Molecular Case Studies: Rapid Communications Rapid whole genome sequencing identifies a homozygous novel variant, His540Arg, in HSD17B4 resulting in D-bifunctional protein deficiency disorder diagnosis. Running title: HSD17B4 His540Arg novel variant Lane Savage ([email protected])1,2*, Stacie D Adams ([email protected])1,3, Kiely James ([email protected])4 , Shimul Chowdhury ([email protected])4 , Surender Rajasekaran ([email protected])1,5,6, Jeremy W. Prokop ([email protected])1,2, Caleb Bupp ([email protected])1,3# 1 Department of Pediatrics and Human Development, College of Human Medicine, Michigan State University, Grand Rapids, MI 49503, USA 2 Department of Pharmacology and Toxicology, Michigan State University, East Lansing MI 48824, USA 3 Spectrum Health/Helen DeVos Children’s Hospital Medical Genetics, Grand Rapids, 49503, MI, USA 4 Rady Children’s Institute for Genomic Medicine, 7910 Frost Street MC5129, San Diego, CA 92123, USA 5 Pediatric Intensive Care Unit, Helen DeVos Children’s Hospital, Grand Rapids, MI, 49503, USA 6 Office of Research, Spectrum Health, 15 Michigan Street NE, Grand Rapids, MI 49503, USA #Corresponding Author: Caleb Bupp ([email protected]) ABSTRACT (234/250): Rapid whole genome sequencing (rWGS) allows for a diagnosis to be made quickly and impact medical management, particularly in critically ill children. Variants identified by this approach are often not identified using other testing methodologies, such as carrier screening or gene sequencing panels, targeted panels, or chromosomal microarrays. However, rWGS can identify variants of uncertain significance (VUS), which challenges clinicians in the rapid return of information to families. -

Role of Catalase in Oxidative Stress-And Age-Associated
Hindawi Oxidative Medicine and Cellular Longevity Volume 2019, Article ID 9613090, 19 pages https://doi.org/10.1155/2019/9613090 Review Article Role of Catalase in Oxidative Stress- and Age-Associated Degenerative Diseases Ankita Nandi,1 Liang-Jun Yan ,2 Chandan Kumar Jana,3 and Nilanjana Das 1 1Department of Biotechnology, Visva-Bharati University, Santiniketan, West Bengal 731235, India 2Department of Pharmaceutical Sciences, UNT System College of Pharmacy, University of North Texas Health Science Center, Fort Worth, TX 76107, USA 3Department of Chemistry, Purash-Kanpur Haridas Nandi Mahavidyalaya, P.O. Kanpur, Howrah, West Bengal 711410, India Correspondence should be addressed to Nilanjana Das; [email protected] Received 25 March 2019; Revised 18 June 2019; Accepted 14 August 2019; Published 11 November 2019 Academic Editor: Cinzia Signorini Copyright © 2019 Ankita Nandi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Reactive species produced in the cell during normal cellular metabolism can chemically react with cellular biomolecules such as nucleic acids, proteins, and lipids, thereby causing their oxidative modifications leading to alterations in their compositions and potential damage to their cellular activities. Fortunately, cells have evolved several antioxidant defense mechanisms (as metabolites, vitamins, and enzymes) to neutralize or mitigate the harmful effect of reactive species and/or their byproducts. Any perturbation in the balance in the level of antioxidants and the reactive species results in a physiological condition called “oxidative stress.” A catalase is one of the crucial antioxidant enzymes that mitigates oxidative stress to a considerable extent by destroying cellular hydrogen peroxide to produce water and oxygen. -

Identification of Expression Qtls Targeting Candidate Genes For
ISSN: 2378-3648 Salleh et al. J Genet Genome Res 2018, 5:035 DOI: 10.23937/2378-3648/1410035 Volume 5 | Issue 1 Journal of Open Access Genetics and Genome Research RESEARCH ARTICLE Identification of Expression QTLs Targeting Candidate Genes for Residual Feed Intake in Dairy Cattle Using Systems Genomics Salleh MS1,2, Mazzoni G2, Nielsen MO1, Løvendahl P3 and Kadarmideen HN2,4* 1Department of Veterinary and Animal Sciences, Faculty of Health and Medical Sciences, University of Copenhagen, Denmark Check for 2Department of Bio and Health Informatics, Technical University of Denmark, Lyngby, Denmark updates 3Department of Molecular Biology and Genetics-Center for Quantitative Genetics and Genomics, Aarhus University, AU Foulum, Tjele, Denmark 4Department of Applied Mathematics and Computer Science, Technical University of Denmark, Lyngby, Denmark *Corresponding author: Kadarmideen HN, Department of Applied Mathematics and Computer Science, Technical University of Denmark, DK-2800, Kgs. Lyngby, Denmark, E-mail: [email protected] Abstract body weight gain and net merit). The eQTLs and biological pathways identified in this study improve our understanding Background: Residual feed intake (RFI) is the difference of the complex biological and genetic mechanisms that de- between actual and predicted feed intake and an important termine FE traits in dairy cattle. The identified eQTLs/genet- factor determining feed efficiency (FE). Recently, 170 can- ic variants can potentially be used in new genomic selection didate genes were associated with RFI, but no expression methods that include biological/functional information on quantitative trait loci (eQTL) mapping has hitherto been per- SNPs. formed on FE related genes in dairy cows. In this study, an integrative systems genetics approach was applied to map Keywords eQTLs in Holstein and Jersey cows fed two different diets to eQTL, RNA-seq, Genotype, Data integration, Systems improve identification of candidate genes for FE. -

Geneseq PLUS
GeneSeq® PLUS Specimen ID: 00000000050 Container ID: H0655 Control ID: Acct #: LCA-BN Phone: SAMPLE REPORT, F-630068 Patient Details Specimen Details Physician Details DOB: 05/05/1995 Date Collected: 08/05/2019 12:00 (Local) Ordering: Age (yyy/mm/dd): 024/07/04 Date Received: 08/06/2019 Referring: Gender: Female Date Entered: 08/06/2019 ID: Patient ID: 00000000050 Date Reported: 08/16/2019 13:21 (Local) NPI: Ethnicity: Unknown Specimen Type: Blood Lab ID: MNEGA Indication: MAN2B1 carrier testing Genetic Counselor: SUMMARY: POSITIVE POSITIVE RESULTS DISORDER (GENE) RESULTS INTERPRETATION Alpha-mannosidosis POSITIVE Predicted to be a carrier. Genetic counseling is (MAN2B1) Heterozygous for c.2248C>T recommended. NMID: NM_000528 (p.Arg750Trp), pathogenic Chr19:12760746 (GRCh37) Risk: AT INCREASED RISK FOR AFFECTED PREGNANCY. See Additional Clinical Information. Genetic counseling is recommended to discuss the potential clinical and/or reproductive implications of positive results, as well as recommendations for testing family members and, when applicable, this individual's partner. Genetic counseling services are available. To access Integrated Genetics Genetic Counselors please visit www.integratedgenetics.com/genetic-counseling or call (855) GC-CALLS (855-422-2557). ADDITIONAL CLINICAL INFORMATION Alpha-mannosidosis: Alpha-mannosidosis is an autosomal recessive lysosomal storage disorder with variable severity and age at onset. Signs and symptoms may include progressive neurological deterioration, intellectual disability, skeletal and facial abnormalities, immunodeficiency, and hearing impairment. Bone marrow transplant may be an option for some individuals. Treatment is otherwise supportive. (Malm, PMID:18651971). If this individual's reproductive partner is also a carrier of a pathogenic variant in the same gene the risk for an affected fetus is 25%. -

Inheritest 500 PLUS
Inheritest® 500 PLUS 525 genes Specimen ID: 00000000010 Container ID: H0651 Control ID: Acct #: LCA-BN Phone: SAMPLE REPORT, F-630049 Patient Details Specimen Details Physician Details DOB: 01/01/1991 Date Collected: 08/05/2019 12:00 (Local) Ordering: Age (yyy/mm/dd): 028/07/04 Date Received: 08/06/2019 Referring: Gender: Female Date Entered: 08/06/2019 ID: Patient ID: 00000000010 Date Reported: 08/21/2019 15:29 (Local) NPI: Ethnicity: Unknown Specimen Type: Blood Lab ID: MNEGA Indication: Carrier screening Genetic Counselor: None SUMMARY: POSITIVE POSITIVE RESULTS DISORDER (GENE) RESULTS INTERPRETATION Spinal muscular atrophy AT RISK AT RISK to be a silent carrier (2+0). For ethnic-specific risk (SMN1) 2 copies of SMN1; positive for revisions see Methods/Limitations. Genetic counseling is NMID: NM_000344 c.*3+80T>G SNP recommended. Risk: AT INCREASED RISK FOR AFFECTED PREGNANCY. See Additional Clinical Information. NEGATIVE RESULTS DISORDER (GENE) RESULTS INTERPRETATION Cystic fibrosis NEGATIVE This result reduces, but does not eliminate the risk to be a (CFTR) carrier. NMID: NM_000492 Risk: NOT at an increased risk for an affected pregnancy. Fragile X syndrome NEGATIVE: Not a carrier of a fragile X expansion. (FMR1) 29 and 36 repeats NMID: NM_002024 Risk: NOT at an increased risk for an affected pregnancy. ALL OTHER DISORDERS NEGATIVE This result reduces, but does not eliminate the risk to be a carrier. Risk: The individual is NOT at an increased risk for having a pregnancy that is affected with one of the disorders covered by this test. For partner's gene-specific risks, visit www.integratedgenetics.com. -

Hamdan Medical Journal 2012; 5:313–326 (
Hamdan Medical Journal 2012; 5:313–326 (http://dx.doi.org/10.7707/hmj.v5i3.212) REVIEW FOR THE SHEIKH HAMDAN BIN RASHID AL MAKTOUM AWARD FOR MEDICAL SCIENCES Clinical, biochemical and genetic aspects of peroxisomal disorders – an expanding group of genetic diseases in humans Ronald JA Wanders University of Amsterdam, Academic Medical Centre, Department of Clinical Chemistry and Paediatrics, Emma Children’s Hospital, Laboratory Genetic Metabolic Diseases, Amsterdam, the Netherlands Abstract clitoris hypertrophy, camptodactyly and simian creases. Unaware of this publication Smith, Opitz and Zellweger syndrome (ZS) in its classic form is an autosomal recessive lethal 2 disease characterized by the absence of morphologically recognizable Inhorn in 1965 described ‘a syndrome of multiple peroxisomes. Detailed studies on ZS in the early 1980s have led to developmental defects including polycystic kidneys the discovery of a set of peroxisomal biomarkers in blood which has and intrahepatic biliary dysgenesis in two sibs’, revolutionized our knowledge about peroxisomes and peroxisomal disorders, who presented with a large number of comparable and formed the basis for the discovery of the group of peroxisomal diseases defects including severe hypotonia, high forehead, known at present. Peroxisomal disorders are classi!ed into two distinct shallow supraorbital ridges, camptodactyly, minor groups including the disorders of peroxisome biogenesis (group 1) and anomalies of the eyes, ears, palate and hands, peroxisome function (group 2). The enzymatic and molecular basis of most and failure to thrive. Two years later, Passarge and peroxisomal disorders has been identi!ed through the years and pre- and McAdams3 described #ve sisters with similar clinical post-natal diagnostic methods have been established. -
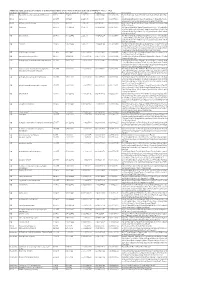
Additional Tables.Xlsx
Additional Table 2 Enriched pathways of downregulated DEGs by the phosphorylation deficient of CRMP2 (P <0.05, q <0.2) Database Description Ratio of DEGs Ratio of universe P -value P adjust q -value Gene name KEGG Biosynthesis of unsaturated fatty acids 8/175 21/4117 1.15E-06 0.000236049 0.000232715 Elovl6/Hsd17b4/Fads2/Elovl4/Scd1/Fads1/Hsd17b12/Hac d3 KEGG Lysosome 15/175 98/4117 1.22E-05 0.00089044 0.000877866 Gla/Man2b1/Entpd4/Gm2a/Acp2/Entpd4/Entpd4b/Scarb 2/Lamp2/Hexa/Asah1/Gns/Lgmn/Smpd1/Ctsd/Ctsb KEGG Fatty acid metabolism 10/175 45/4117 1.30E-05 0.00089044 0.000877866 Elovl6/Acsl1/Hsd17b4/Acaa2/Fads2/Elovl4/Scd1/Fads1/Hs d17b12/Hacd3 GO lysosome 29/455 304/11442 1.14E-05 0.002429124 0.002071406 Gla/Rptor/Man2b1/Gm2a/Acp2/Psen1/Slc35f6/Scarb2/Ba ce1/Snx14/Rnaset2a/Ncstn/Lamp2/Hexa/Plbd2/Asah1/Gn s/Ostm1/Ank3/Pon2/Mlc1/Gja1/Lgmn/Smpd1/Rnf13/Arl8 b/Ctsd/Ctsb/Rab7 GO lytic vacuole 29/455 305/11442 1.21E-05 0.002429124 0.002071406 Gla/Rptor/Man2b1/Gm2a/Acp2/Psen1/Slc35f6/Scarb2/Ba ce1/Snx14/Rnaset2a/Ncstn/Lamp2/Hexa/Plbd2/Asah1/Gn s/Ostm1/Ank3/Pon2/Mlc1/Gja1/Lgmn/Smpd1/Rnf13/Arl8 b/Ctsd/Ctsb/Rab7 GO vacuole 33/455 374/11442 1.47E-05 0.002429124 0.002071406 Gla/Rptor/Abcb6/Man2b1/Entpd4/Gm2a/Acp2/Psen1/Ent pd4b/Slc35f6/Scarb2/Bace1/Snx14/Rnaset2a/Ncstn/Lamp 2/Hexa/Plbd2/Asah1/Gns/Ostm1/Ank3/Pon2/Atg12/Mlc1 /Gja1/Lgmn/Smpd1/Rnf13/Arl8b/Ctsd/Ctsb/Rab7 GO GABA-ergic synapse 12/455 88/11442 0.000178214 0.020677491 0.017632481 Gabrb1/Slitrk3/Iqsec3/Gabrb2/Nlgn3/Gucy1a1/Rims2/Cd h13/Clstn3/Sv2a/Pak1/Atp2b1 GO neuromuscular junction 11/455