1 Single Cell Derived Organoids Capture the Self-Renewing Subpopulations of Metastatic Ovarian Cancer Tania Velletri1&, Eman
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Product Information
Product information NANOG 1-154aa Human, His-tagged, Recombinant, E.coli Cat. No. IBATGP1124 Full name: Homeobox protein NANOG NCBI Accession No.: NP_079141 Synonyms: Homeobox transcription factor Nanog, homeobox transcription factor Nanog-delta 48 Description: NANOG, also known as nanog homeobox, is a member of the homeobox family of DNA binding transcription factors that has been shown to maintain pluripotency of embryonic stem cells. Nanog expression counteracts the differentiation-promoting signals induced by the extrinsic factors LIF, Stat3 and BMP. Once NANOG expression is down-regulated, cell differentiation can proceed. Recombinant human NANOG protein, fused to His- tag at N-terminus, was expressed in E.coli and purified by using conventional chromatography. Form: Liquid. 20mM Tris-HCl buffer (pH8.0) containing 20% glycerol, 1mM DTT Molecular Weight: 19.6 kDa (174aa), confirmed by MALDI-TOF (Molecular weight on SDS-PAGE will appear higher) Purity: > 90% by SDS - PAGE Concentration: 0.25 mg/ml (determined by Bradford assay) 15% SDS-PAGE (3ug) Sequences of amino acids: MGSSHHHHHH SSGLVPRGSH MSVDPACPQS LPCFEASDCK ESSPMPVICG PEENYPSLQM SSAEMPHTET VSPLPSSMDL LIQDSPDSST SPKGKQPTSA EKSVAKKEDK VPVKKQKTRT VFSSTQLCVL NDRFQRQKYL SLQQMQELSN ILNLSYKQVK TWFQNQRMKS KRWQ General references: Clark A T., et al. (2004) Stem Cells. 22:169-179. Chambers I., et al. (2003) Cell Res. 13:499-502. Storage: Can be stored at +4°C short term (1-2 weeks). For long term storage, aliquot and store at -20°C or -70°C. Avoid repeated freezing and thawing cycles. For research use only. This product is not intended or approved for human, diagnostics or veterinary use. Immuno-Biological Laboratories, Inc. (IBL-America) 8201 Central Ave. -

Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation
From THE DEPARTMENT OF MICROBIOLOGY TUMOR AND CELL BIOLOGY (MTC) Karolinska Institutet, Stockholm, Sweden ROLE OF MITOCHONDRIAL RIBOSOMAL PROTEIN S18-2 IN CANCEROGENESIS AND IN REGULATION OF STEMNESS AND DIFFERENTIATION Muhammad Mushtaq Stockholm 2017 All previously published papers were reproduced with permission from the publisher. Published by Karolinska Institutet. Printed by E-Print AB 2017 © Muhammad Mushtaq, 2017 ISBN 978-91-7676-697-2 Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation THESIS FOR DOCTORAL DEGREE (Ph.D.) By Muhammad Mushtaq Principal Supervisor: Faculty Opponent: Associate Professor Elena Kashuba Professor Pramod Kumar Srivastava Karolinska Institutet University of Connecticut Department of Microbiology Tumor and Cell Center for Immunotherapy of Cancer and Biology (MTC) Infectious Diseases Co-supervisor(s): Examination Board: Professor Sonia Lain Professor Ola Söderberg Karolinska Institutet Uppsala University Department of Microbiology Tumor and Cell Department of Immunology, Genetics and Biology (MTC) Pathology (IGP) Professor George Klein Professor Boris Zhivotovsky Karolinska Institutet Karolinska Institutet Department of Microbiology Tumor and Cell Institute of Environmental Medicine (IMM) Biology (MTC) Professor Lars-Gunnar Larsson Karolinska Institutet Department of Microbiology Tumor and Cell Biology (MTC) Dedicated to my parents ABSTRACT Mitochondria carry their own ribosomes (mitoribosomes) for the translation of mRNA encoded by mitochondrial DNA. The architecture of mitoribosomes is mainly composed of mitochondrial ribosomal proteins (MRPs), which are encoded by nuclear genomic DNA. Emerging experimental evidences reveal that several MRPs are multifunctional and they exhibit important extra-mitochondrial functions, such as involvement in apoptosis, protein biosynthesis and signal transduction. Dysregulations of the MRPs are associated with severe pathological conditions, including cancer. -

Cell Cycle and Pluripotency: Convergence on Octamer‑Binding Transcription Factor 4 (Review)
MOLECULAR MEDICINE REPORTS 16: 6459-6466, 2017 Cell cycle and pluripotency: Convergence on octamer‑binding transcription factor 4 (Review) SHIQI SHE1*, QUCHENG WEI2*, BO KANG1 and YING-JIE WANG1 1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang 310003; 2Cardiovascular Key Lab of Zhejiang, Department of Cardiology, The Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang 310009, P.R. China Received December 17, 2016; Accepted July 14, 2017 DOI: 10.3892/mmr.2017.7489 Abstract. Embryonic stem cells (ESCs) have unlimited expan- 1. Introduction sion potential and the ability to differentiate into all somatic cell types for regenerative medicine and disease model studies. Embryonic stem cells (ESCs) are characterized by unlimited Octamer-binding transcription factor 4 (OCT4), encoded by proliferation (self-renewal) and the ability to differentiate into the POU domain, class 5, transcription factor 1 gene, is a tran- three primary germ layers, namely the endoderm, mesoderm scription factor vital for maintaining ESC pluripotency and and ectoderm (pluripotency) (1-4). It has been established somatic reprogramming. Many studies have established that that complicated regulatory networks are present in ESCs the cell cycle of ESCs is featured with an abbreviated G1 phase that critically maintain the state of self-renewal and pluri- and a prolonged S phase. Changes in cell cycle dynamics are potency for later development (5,6). Several transcription intimately associated with the state of ESC pluripotency, and factors (TFs), including octamer-binding transcription factor manipulating cell-cycle regulators could enable a controlled 4 (OCT4), SRY-box 2 (SOX2) and homeobox protein NANOG differentiation of ESCs. -

Studies Relating to the Differentiation of Human Embryonic Stem Cells
Studies relating to the differentiation of human embryonic stem cells A thesis submitted to the University of Manchester for the degree of Doctor of Philosophy in the Faculty of Life Sciences 2015 Georgios Anyfantis List of Figures .................................................................................................... 10 List of Tables ...................................................................................................... 13 Declaration ........................................................................................................ 15 Copyright statement .......................................................................................... 15 Acknowledgements ........................................................................................... 16 Abbreviations .................................................................................................... 17 Chapter 1: Introduction ...................................................................................... 23 1.1 Diabetes Mellitus ......................................................................................... 23 1.2 Pancreatic function and Organogenesis ........................................................ 24 1.2.1 Pancreatic Function .................................................................................... 24 1.2.2 Pancreatic Organogenesis ........................................................................... 26 1.3 Signalling Pathways associated with pancreas development ....................... -

Human Cell Modeling for Cardiovascular Diseases
International Journal of Molecular Sciences Review Human Cell Modeling for Cardiovascular Diseases Melania Lippi 1, Ilaria Stadiotti 1, Giulio Pompilio 1,2 and Elena Sommariva 1,* 1 Unit of Vascular Biology and Regenerative Medicine, Centro Cardiologico Monzino IRCCS, 20138 Milan, Italy; [email protected] (M.L.); [email protected] (I.S.); [email protected] (G.P.) 2 Department of Clinical Sciences and Community Health, Università degli Studi di Milano, 20122 Milan, Italy * Correspondence: [email protected]; Tel.: +39-02-58002026; Fax: +39-02-58002342 Received: 13 August 2020; Accepted: 31 August 2020; Published: 2 September 2020 Abstract: The availability of appropriate and reliable in vitro cell models recapitulating human cardiovascular diseases has been the aim of numerous researchers, in order to retrace pathologic phenotypes, elucidate molecular mechanisms, and discover therapies using simple and reproducible techniques. In the past years, several human cell types have been utilized for these goals, including heterologous systems, cardiovascular and non-cardiovascular primary cells, and embryonic stem cells. The introduction of induced pluripotent stem cells and their differentiation potential brought new prospects for large-scale cardiovascular experiments, bypassing ethical concerns of embryonic stem cells and providing an advanced tool for disease modeling, diagnosis, and therapy. Each model has its advantages and disadvantages in terms of accessibility, maintenance, throughput, physiological relevance, recapitulation of the disease. A higher level of complexity in diseases modeling has been achieved with multicellular co-cultures. Furthermore, the important progresses reached by bioengineering during the last years, together with the opportunities given by pluripotent stem cells, have allowed the generation of increasingly advanced in vitro three-dimensional tissue-like constructs mimicking in vivo physiology. -

Gene Expression Profiling in Peripheral Blood Nuclear Cells in Patients with Refractory Ischaemic End-Stage Heart Failure
J Appl Genet 51(3), 2010, pp. 353–368 Original article Gene expression profiling in peripheral blood nuclear cells in patients with refractory ischaemic end-stage heart failure S. Szmit1,2, M. Jank3, H. Maciejewski4, M. Grabowski1, R. Glowczynska1, A. Majewska3, K. J. Filipiak1, T. Motyl3, G. Opolski1 1First Department of Cardiology, Medical University of Warsaw, Poland 2Department of Oncology, Military Institute of Medicine, Warsaw, Poland 3Department of Physiological Sciences, Faculty of Veterinary Medicine, Warsaw University of Life Sciences, Warsaw, Poland 4Institute of Computer Engineering, Control and Robotics (I-6), Wroc³aw University of Technology, Wroc³aw, Poland Abstract. Functional analysis of up- and down-regulated genes might reveal whether peripheral blood cells may be considered as a material of diagnostic or prognostic value in patients with end-stage heart failure (HF). The aim of the present study was to compare the transcriptomic profile of peripheral blood nuclear cells from 6 male patients with ischaemic end-stage HF with those of 6 male patients with asymptomatic cardiac dysfunction. The expression of genes in peripheral blood nuclear cells in both groups of patients was measured using whole-genome oligonucleotide microarrays utilizing 35 035 oligonucleotide probes. Microarray analyses re- vealed 130 down-regulated genes and 15 up-regulated genes in the patients with end-stage HF. Some of the down-regulated genes belonged to the pathways that other studies have shown to be down-regulated in cardiomyopathy. We also identified up-regulated genes that have been correlated with HF severity (CXCL16) and genes involved in the regulation of expression of platelet activation factor receptor (PTAFR, RBPSUH, MCC, and PSMA7). -
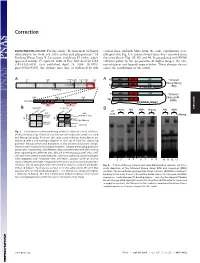
Requirement of Nanog Dimerization for Stem Cell Self-Renewal and Pluripotency
Correction DEVELOPMENTAL BIOLOGY. For the article ‘‘Requirement of Nanog vertical lines, multiple blots from the same experiments were dimerization for stem cell self-renewal and pluripotency,’’ by juxtaposed in Fig. 1A, and irrelevant lanes were removed from Jianlong Wang, Dana N. Levasseur, and Stuart H. Orkin, which the same blot in Figs. 1B,3D, and 4B. In accordance with PNAS appeared in issue 17, April 29, 2008, of Proc Natl Acad Sci USA editorial policy on the preparation of digital images, the cor- (105:6326–6331; first published April 24, 2008; 10.1073͞ rected figures and legends appear below. These changes do not pnas.0802288105), the authors note that, as indicated by thin affect the conclusions of the article. Void ND HD CD A 4MD 700kD 440kD 232kD 158kD 63kD A N Tethered ND HD CD Nanog-Nanog N (NN) GT(GGGS)4GGGT Anti-Nanog ND HD CD N Tethered 100kD 63kD 30kD Nanog-NH NH ND HD Anti-Nanog (NNH) GT(GGGS)4GGGT dimer monomer B NanogV5 FL-Nanog B CORRECTION or og V5 Vector NN NNH Nanog NN NNH ect an V5 og wt -V -N WB: irA an FL FL B N WB: COS αNanog (Anti-V5) <n.s. (Anti-M2 FLAG) -+ -+ -+-+ -+-+ <NanogV5 * <FL-Nanog IP: IP: <NanogV5 FLAG <FL-Nanog Anti-V5 Anti- M2 * (Anti-Nanog) (Anti-V5) < < < < <NanogV5 < NanogV5 < (Anti-V5) Input Input < BirAV5 Fig. 1. Confirmation of Nanog–Nanog interaction (dimerization). (A) West- * * * * * ern blot analysis of gel filtration fractions containing protein complexes using * anti-Nanog antibody. Fractions that may contain Nanog homodimers are indicated with a red rectangle (Upper). -

The Role of the Cytoskeleton in Pluripotent Stem Cell Differentiation
THE ROLE OF THE CYTOSKELETON IN PLURIPOTENT STEM CELL DIFFERENTIATION AN ABSTRACT SUBMITTED ON THE 21ST DAY OF AUGUST, 2013 TO THE DEPARTMENT OF BIOMEDICAL ENGINEERING OF THE SCHOOL OF SCIENCE AND ENGINEERING AT TULANE UNIVERSITY IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY BY ___________________________ EMMA T. PINEDA FORTIN APPROVED: ___________________________ TABASSUM AHSAN, Ph.D. (COMMITTEE CHAIR) __________________________ WALTER LEE MURFEE, Ph.D. __________________________ DAMIR KHISMATULLIN, Ph.D. ABSTRACT An understanding of the pathways responsible for differentiation in pluripotent stem cells (PSCs) would accelerate their translation to medical therapies. Specifically, studies that identify criteria for the better design of experiments targeting certain phenotypes would allow for the generation of cell sources adequate for transplantation. In this dissertation, we aimed at elucidating the role of the cytoskeleton in the spontaneous differentiation of PSCs in two dimensional (2D) and three dimensional (3D) microenvironments. First, we quantified the expression of the cytoskeleton in ESCs, iPSCs, and the iPSC source phenotype, showing that there were indeed differences in the expression of microfilaments and certain intermediate filaments among all three phenotypes. Next, we found that there were inherent differences in ESC differentiation when cultured in 2D and 3D microenvironments. Lastly, alterations in the cytoskeleton were found to decrease mesodermal differentiation in 3D culture, while -
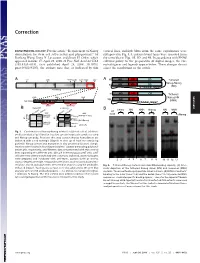
Requirement of Nanog Dimerization for Stem Cell Self-Renewal and Pluripotency
Correction DEVELOPMENTAL BIOLOGY. For the article ‘‘Requirement of Nanog vertical lines, multiple blots from the same experiments were dimerization for stem cell self-renewal and pluripotency,’’ by juxtaposed in Fig. 1A, and irrelevant lanes were removed from Jianlong Wang, Dana N. Levasseur, and Stuart H. Orkin, which the same blot in Figs. 1B,3D, and 4B. In accordance with PNAS appeared in issue 17, April 29, 2008, of Proc Natl Acad Sci USA editorial policy on the preparation of digital images, the cor- (105:6326–6331; first published April 24, 2008; 10.1073͞ rected figures and legends appear below. These changes do not pnas.0802288105), the authors note that, as indicated by thin affect the conclusions of the article. Void ND HD CD A 4MD 700kD 440kD 232kD 158kD 63kD A N Tethered ND HD CD Nanog-Nanog N (NN) GT(GGGS)4GGGT Anti-Nanog ND HD CD N Tethered 100kD 63kD 30kD Nanog-NH NH ND HD Anti-Nanog (NNH) GT(GGGS)4GGGT dimer monomer B NanogV5 FL-Nanog B CORRECTION or og V5 Vector NN NNH Nanog NN NNH ect an V5 og wt -V -N WB: irA an FL FL B N WB: COS αNanog (Anti-V5) <n.s. (Anti-M2 FLAG) -+ -+ -+-+ -+-+ <NanogV5 * <FL-Nanog IP: IP: <NanogV5 FLAG <FL-Nanog Anti-V5 Anti- M2 * (Anti-Nanog) (Anti-V5) < < < < <NanogV5 < NanogV5 < (Anti-V5) Input Input < BirAV5 Fig. 1. Confirmation of Nanog–Nanog interaction (dimerization). (A) West- * * * * * ern blot analysis of gel filtration fractions containing protein complexes using * anti-Nanog antibody. Fractions that may contain Nanog homodimers are indicated with a red rectangle (Upper). -

Gli Affects the Stemness and Prognosis of Epithelial Ovarian Cancer Via Homeobox Protein NANOG
MOLECULAR MEDICINE REPORTS 23: 128, 2021 Gli affects the stemness and prognosis of epithelial ovarian cancer via homeobox protein NANOG HUAN ZHAO1,2, NA LI3, YICUN PANG1, JUN ZHAO1 and XIAOHUA WU2,4 1Department of Obstetrics and Gynecology, The Third Hospital of Hebei Medical University; 2Teaching and Research Section of Obstetrics and Gynecology, Hebei Medical University; 3Department of Oncology, Hebei General Hospital; 4Department of Obstetrics and Gynecology, The Fourth Hospital of Shijiazhuang, Shijiazhuang, Hebei 050000, P.R. China Received July 18, 2020; Accepted November 10, 2020 DOI: 10.3892/mmr.2020.11767 Abstract. Gli proteins are key transcription factors of the Introduction Hedgehog (HH) signaling pathway, which is associated with tumorigenesis and drug resistance. However, the role of the Ovarian cancer is a malignant tumor of the female reproduc‑ HH signaling pathway in epithelial ovarian cancer (EOC) tive system characterized by late detection, high risk of drug remains unclear. Studies have demonstrated that in some resistance, and low 5‑year survival rate (1). It was previously tumors, homeobox protein NANOG (NANOG), a known stem reported that the mechanisms underlying the morbidity, cell marker, is a downstream effector of Gli. However, limited metastasis, drug resistance and recurrence of ovarian cancer research has been conducted on the association between Gli are complex processes involving multiple factors (2). One and NANOG in EOC, particularly regarding their roles in the reason for the poor prognosis of ovarian cancer is the lack tumor stemness, such as tumor development, drug resistance of early diagnosis and prognosis markers (2). In addition, the and patient prognosis. Thus, the aim of the present study was emergence of chemoresistance in ovarian cancer is a major to explore the aforementioned issues. -

82419926.Pdf
Digestive and Liver Disease 47 (2015) 826–835 View metadata, citation and similar papers at core.ac.uk brought to you by CORE Contents lists available at ScienceDirect provided by Elsevier - Publisher Connector Digestive and Liver Disease journal homepage: www.elsevier.com/locate/dld Review Article Yes-associated protein in the liver: Regulation of hepatic development, repair, cell fate determination and tumorigenesis a d a,b,c a,b,c,∗ Quy Nguyen , Robert A. Anders , Gianfranco Alpini , Haibo Bai a Research, Central Texas Veterans Health Care System, Temple, TX, United States b Digestive Diseases Research Center, BaylorScott&White Healthcare, Temple, TX, United States c Department of Internal Medicine and Medical Physiology, Texas A&M Health Science Center, Temple, TX, United States d Department of Pathology, Johns Hopkins University, Baltimore, MD, United States a r t i c l e i n f o a b s t r a c t Article history: The liver is a vital organ that plays a major role in many bodily functions from protein production and Received 17 March 2015 blood clotting to cholesterol, glucose and iron metabolism and nutrition storage. Maintenance of liver Accepted 14 May 2015 homeostasis is critical for these essential bodily functions and disruption of liver homeostasis causes Available online 22 May 2015 various kinds of liver diseases, some of which have high mortality rate. Recent research advances of the Hippo signalling pathway have revealed its nuclear effector, Yes-associated protein, as an important Keywords: regulator of liver development, repair, cell fate determination and tumorigenesis. Therefore, a precise Hippo signalling pathway control of Yes-associated protein activity is critical for the maintenance of liver homeostasis. -

Epstein-Barr Virus Induction of the Hedgehog Signalling Pathway Imposes a Stem Cell-Like Phenotype on Human Epithelial Cells –
Epstein-Barr Virus Induction of the Hedgehog Signalling Pathway Imposes a Stem Cell-Like Phenotype on Human Epithelial Cells – Implications for the Pathogenesis of Nasopharyngeal Carcinoma by REBECCA J. PORT A thesis submitted to the University of Birmingham for the degree of DOCTOR OF PHILOSOPHY School of Cancer Sciences College of Medical and Dental Sciences University of Birmingham September 2013 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Nasopharyngeal carcinoma (NPC) is endemic in Southern China and South East Asia, causally linked to Epstein-Barr virus (EBV) infection, and frequently shows dysregulation in a number of stem cell maintenance signalling pathways. This thesis has endeavoured to investigate the status of one of these pathways; the Hedgehog (HH) signalling pathway, in NPC tumours, and reveals the novel finding that EBV is able to active the HH signalling pathway through autocrine induction of the SHH ligand in the C666.1 authentic EBV-positive NPC-derived cell line and latently infected epithelial carcinoma cell lines. This study demonstrates that constitutive engagement of the HH pathway in EBV-infected epithelial cells in vitro induces the expression of a number of stemness-associated genes and imposes stem-like characteristics.