Foi Vehicles
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Il Cristianesimo in Egitto Luci E Ombre in Abydos La Tomba
egittologia.net magazine in questo numero: IL CRISTIANESIMO IN EGITTO EGITTO A VENEZIA LUCI E OMBRE IN ABYDOS SPECIALE NEFERTARI LA TOMBA QV66 AREA ARCHEOLOGICA TEBANA IL VILLAGGIO DI DEIR EL-MEDINA EGITTO IN PILLOLE ISCRIZIONI IERATICHE NELLA TOMBA DI THUTMOSI IV Italiani in Egitto: Ernesto Schiaparelli | L’Arte di Shamira | I papiri di Carla BOLLETTINO INFORMATIVO DELL'ASSOCIAZIONE EGITTOLOGIA.NET NUMERO 3 e d i t o r i a l e La prolungata e precoce presenza di questo Confesso che questo numero di EM – Egitto- insolito e intenso caldo, dà l’impressione che logia.net Magazine è stato sul punto di non l’estate stia già volgendo al termine, anche se uscire! La prossimità con il ferragosto e il in realtà la legna accumulata per l’inverno caldo scoraggiante, soprattutto nelle due set- dovrà aspettare ancora molto tempo prima di timane centrali del mese di luglio – periodo in essere utile. cui il terzo numero del magazine ha comin- Curioso come hanno deciso di chiamare le tre ciato a prendere vita – ci avevano fatto propen- fasi più intense del caldo i meteorologici: Sci- dere per una sospensione, procrastinandone pione, Caronte e Minosse. Curioso perché mi l’uscita direttamente a ottobre. vien da pensare che l’epiteto “Africano” di Sci- Ma abbiamo resistito alla tentazione, sospen- pione e il collegamento con l’Ade che è possi- dendo solo una parte dei temi che abbiamo bile fare con Caronte e Minosse, abbia cominciato a trattare nei numeri precedenti, richiamato alla mente degli scienziati il con- come ci è stato richiesto dagli autori degli cetto di “caldo”. -

Connect Series Accessories
Connect Series Accessories Our energy working for y ou.™ www.cumminspower.com 2013 Cummins Power Generation Inc. All rights reserved. Specifications are subject to change without notice. Cummins Power Generation and Cummins are registered trademarks of Cummins Inc. “Our energy working for you.” is a trademark of Cummins Power Generation. Other company, product, or service names may be trademarks or service marks of others. NAACSB-6026-EN (03/15) CONNECT SERIES ACCESSORIES Table of Contents DISPLAYS .................................................................................................................................................................. 1 HMI 211 Remote Unit (0541-1394) .................................................................................................... 1 RS In-Home Display (A046K103)....................................................................................................... 2 HMI 220 Remote Unit (A041J582) ................................................................................................... 3 ANNUNCIATORS (HMI 113 UNIT) ........................................................................................................................... 4 Annunciator – RS485 Panel Mount (A045J199) ................................................................................. 4 Annunciator – RS485 Panel Mount w/ Enclosure (A045J201) ............................................................ 4 AUXILIARY ............................................................................................................................................................... -
DSFRA IKEN Report Template
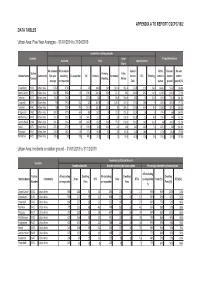
APPENDIX A TO REPORT CSCPC/19/2 DATA TABLES Urban Area: Five-Year Averages – 01/04/2014 to 31/04/2019 Incidents on station grounds Location False Pump Attendances Overview Fires Special Service Alarm All incidents All incidents Special All by On own On own Station Primary: False Station Name Community five-year excluding Co-responder All Primary Secondary Service RTC Flooding station's station station Number Dwelling Alarms average co-responder Calls pumps ground ground (%) Greenbank KV50 Urban Area 878.6 878.6 0 245 104.6 56.6 140.4 361.4 271.8 21.6 24.6 1424.8 974.2 68.4% Danes Castle KV32 Urban Area 832.6 830.8 1.8 198.8 126.4 56.6 72.4 385 248.4 29.2 14.8 1090.6 849.4 77.9% Torquay KV17 Urban Area 744.8 744.8 0 207.8 111 59 96.8 306.8 230 36 15.8 919.8 776.4 84.4% Crownhill KV49 Urban Area 742 741.8 0.2 227 100.6 43 126.4 337.4 177.4 28.6 9 878.4 680.6 77.5% Taunton KV61 Urban Area 734 733.4 0.6 227.8 132.8 56.6 95 284.6 221.6 65.4 8.4 1038.8 901.8 86.8% Bridgwater KV62 Urban Area 584.2 577.6 6.6 160 88.2 38 71.8 231.8 192.4 56 8 774.4 666 86.0% Middlemoor KV59 Urban Area 537.6 535.8 1.8 144.2 91.2 33 53 239.6 153.8 51 8.8 724.4 444 61.3% Camels Head KV48 Urban Area 491.6 491.2 0.4 162.8 85.2 50.4 77.6 178.6 150.2 16.6 11.8 638 390.2 61.2% Yeovil KV73 Urban Area 471.6 471.6 0 139.6 78.6 34.8 61 191 141 46.8 7.4 674.2 569 84.4% Plympton KV47 Urban Area 218.4 204.4 14 57.8 34.8 12 23 87.8 72.4 18.6 3 170.6 135.8 79.6% Plymstock KV51 Urban Area 185.8 185 0.8 48.4 27.4 12 21 76.8 60.6 12.6 2.6 165.4 123.8 74.8% Urban Area: Incidents on -

Expanding the Toolkit for Metabolic Engineering
Expanding the Toolkit for Metabolic Engineering Yao Zong (Andy) Ng Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2016 © 2016 Yao Zong (Andy) Ng All rights reserved ABSTRACT Expanding the Toolkit for Metabolic Engineering Yao Zong (Andy) Ng The essence of metabolic engineering is the modification of microbes for the overproduction of useful compounds. These cellular factories are increasingly recognized as an environmentally-friendly and cost-effective way to convert inexpensive and renewable feedstocks into products, compared to traditional chemical synthesis from petrochemicals. The products span the spectrum of specialty, fine or bulk chemicals, with uses such as pharmaceuticals, nutraceuticals, flavors and fragrances, agrochemicals, biofuels and building blocks for other compounds. However, the process of metabolic engineering can be long and expensive, primarily due to technological hurdles, our incomplete understanding of biology, as well as redundancies and limitations built into the natural program of living cells. Combinatorial or directed evolution approaches can enable us to make progress even without a full understanding of the cell, and can also lead to the discovery of new knowledge. This thesis is focused on addressing the technological bottlenecks in the directed evolution cycle, specifically de novo DNA assembly to generate strain libraries and small molecule product screens and selections. In Chapter 1, we begin by examining the origins of the field of metabolic engineering. We review the classic “design–build–test–analyze” (DBTA) metabolic engineering cycle and the different strategies that have been employed to engineer cell metabolism, namely constructive and inverse metabolic engineering. -

Response to Request for Information
[NOT PROTECTIVELY MARKED] Response to Request for Information Reference FOI 003352 Date 13 March 2019 Hackney Carriage/Private Hire Request: I would like to make a request under the freedom of information act for the following information: Motor Vehicles registered for public hire i.e. Hackney Carriage or Private Hire relating to the period 1st November 2018 to 28th February 2019. Specifically, I would like to know: (If any of these elements are not available, please supply the ones that are) • Vehicle registration • Manufacturer (Make) • Model • Date at which they were first licensed • Date at which the license ceased In response to your request, please find below our response: Reg No. -

ABSTRACT Carl Nicholas Reeves STUDIES in the ARCHAEOLOGY
ABSTRACT Carl Nicholas Reeves STUDIES IN THE ARCHAEOLOGY OF THE VALLEY OF THE KINGS, with particular reference to tomb robbery and the caching of the royal mummies This study considers the physical evidence for tomb robbery on the Theban west bank, and its resultant effects, during the New Kingdom and Third Intermediate Period. Each tomb and deposit known from the Valley of the Kings is examined in detail, with the aims of establishing the archaeological context of each find and, wherever possible, isolating and comparing the evidence for post-interment activity. The archaeological and documentary evidence pertaining to the royal caches from Deir el-Bahri, the tomb of Amenophis II and elsewhere is drawn together, and from an analysis of this material it is possible to suggest the routes by which the mummies arrived at their final destinations. Large-scale tomb robbery is shown to have been a relatively uncommon phenomenon, confined to periods of political and economic instability. The caching of the royal mummies may be seen as a direct consequence of the tomb robberies of the late New Kingdom and the subsequent abandonment of the necropolis by Ramesses XI. Associated with the evacuation of the Valley tombs may be discerned an official dismantling of the burials and a re-absorption into the economy of the precious commodities there interred. STUDIES IN THE ARCHAEOLOGY OF THE VALLEY OF THE KINGS, with particular reference to tomb robbery and the caching of the royal mummies (Volumes I—II) Volume I: Text by Carl Nicholas Reeves Thesis submitted for the degree of Doctor of Philosophy School of Oriental Studies University of Durham 1984 The copyright of this thesis rests with the author. -

The University of Basel Kings Valley Project Finds
The University of Basel Kings Valley Project Finds A New Tomb in the VOK : KV64 by Susanne Bickel & Elina Paulin-Grothe Photos © The University of Basel Kings’ Valley Project he first pharaohs who chose the Valley of the Kings as their burial place still adhered to the tradition of having members of their families and pro- fessional entourages interred in the vicinities of their own tombs. It is only after the Amarna period that the Valley became an (almost) exclusively royal necropolis. The numerous non-royal tombs there have received very T little scientific attention until recently.1 Kmt 18 View of the interior of KV64, with the incribed wooden coffin of the tomb’s occupant, Nehemes-Bastet, a small fu- nerary stela at its foot end. To be granted a tomb in this particular location was certainly considered an important privilege which, as a counterpart, implied some conditions and re- strictions. The most striking rule was the fact that non-royal tombs in the Kings’ Valley were not allowed any wall decoration, quite in contrast to the wonder- fully engraved and painted tomb chapels and burial chambers of the same social groups in other parts of the Theban west bank. Whereas the architectural layout 19 Kmt View of the side wadi of the Valley of the Kings where are located a number of unin- scribed 18th Dynasty tombs, as indicated by the inset graphic. The modern stairs leading up to KVs 34 (Tomb of Thutmose III) & 33 are at the far right of the photo. of royal tombs seems to have followed a regular scheme of drew a map of the area and mentioned certain features of evolution, each plan building upon and enlarging the one the side valley in 1886; Victor Loret discovered the Tomb of its predecessor, non-royal tombs show a wide variety of of Thutmose III (KV34) in 1898; and Howard Carter architectural designs. -

KERRVILLE, TEXAS Om Ed V O Ont Is Set Mage) Igital TBD)
CITY OF KERRVILLE INITIAL DESIGN & PLAN CIVIC WAYFINDING PROGRAM DESIGN INTENT SIGN LOCATION PLAN DRAFTED & THE CITY OF SUBMITTED BY: KERRVILLE, TEXAS 1 3’ VEH.01 Peterson K E R 6’ 2 K E R Vehicular Directional (Lamp Image) Plaza 7 Public 1. Aluminum sign face fabricated from .080 aluminum sheeting painted to 3’ V I L Parking V I L 4’ 9 match City approved colors. Digital Downtown 3 graphics applied to sign face. 8 E 6’ City Hall E 2. Directional arrow cut out of .080 Courthouse aluminum and mounted to sign face 1 using hi-mod adhesive. vinyl and applied to sign face. Font is set to Clearview Highway 2-W. 4. 3.5” Square concrete post painted to match weathered cedar post. 7’ 5. TXDOT approved breakaway base with drilled foundation. 6 6. Decorative metal strapping painted or powdercoated to match City approved colors. 7. Digitally printed graphic applied as background image. 4 8. Aluminum sleeve fabricated to house the sign post. 5 9. Fasteners (color and type TBD). 1 VEH.01 - Front Elevation 2 VEH.01 - Back Elevation Scale: 1” = 1’ Scale: 1” = 1’ 2 VEH.01 2 V ehicular Directional (Lamp Image) 1 1. Aluminum sign face fabricated from .080 aluminum sheeting painted to 4’ match City approved colors. Digital 8 graphics applied to sign face. 1 2. Directional arrow cut out of .080 9 aluminum and mounted to sign face using hi-mod adhesive. 8 3’ 5 vinyl and applied to sign face. Font is set to Clearview Highway 2-W. 4. 3.5” Square concrete post painted to match weathered cedar post. -
A Királyok Völgye Térképes Honlapjának Továbbfejlesztése
A projekt az Európai Unió támogatásával és az Európai Szociális Alap társfinanszírozásával valósul meg (támogatási szerződés száma TÁMOP 4.2.1/B-09/1/KMR-2010-0003) Eötvös Loránd Tudományegyetem Informatikai Kar Térképtudományi és Geoinformatikai Tanszék A Királyok völgye térképes honlapjának továbbfejlesztése Szepesi Petra térképész szakos hallgató Témavezető: José Jesús Reyes Nuñez egyetemi docens Budapest, 2012 Tartalom 1. A szakdolgozat témája - újabb célok ................................................................... 4 2. A téma rövid áttekintése ....................................................................................... 5 3. A korábbi szakdolgozat munkarészei .................................................................. 8 3.1. A térképek .................................................................................................................... 8 3.2. Honlap ......................................................................................................................... 9 4. Újítások ................................................................................................................. 10 4.1. Felhasznált adatok és tartalmak ............................................................................ 10 4.1.1. Leíró adatok ........................................................................................................ 10 4.1.2. Multimédiás adatok ............................................................................................ 10 4.2. A használt szoftverek.............................................................................................. -

Connect Series Accessories
Connect Series Accessories power.cummins.com ©2018 Cummins Inc. | NAAC-6260E-EN (01/18) CONNECT SERIES ACCESSORIES Table of Contents ALTERNATOR HEATERS ................................................................................................................................. 1 ANNUNCIATORS .............................................................................................................................................. 2 AUXILIARY ........................................................................................................................................................ 3 AUXILIARY CONFIGURABLE SIGNAL INPUTS / RELAY OUTPUTS ....................................................................... 3 AUXILIARY OUTPUTS RELAYS ................................................................................................................................ 3 BATTERIES ....................................................................................................................................................... 4 BATTERY ACCESSORIES ............................................................................................................................... 4 BATTERY HEATER KITS .......................................................................................................................................... 4 BATTERY TRAY KITS ............................................................................................................................................... 4 BATTERY CHARGERS .................................................................................................................................... -

Kirksville City Council Newsletter
KIRKSVILLE CITY COUNCIL NEWSLETTER . Mari E. Macomber, City Manager April 29, 2015 SUBJECTS: KIRKSVILLE CELEBRATED ARBOR DAY MML LEGISLATIVE UPDATE BROOK DRIVE CROSSING OVER BEAR CREEK COUNCIL NETBOOK REPLACEMENT OPTIONS ECONOMIC DEVELOPMENT PROJECT UPDATE PURSUING ADVERSE PARTIES FOR DAMAGES FOUNDERS DAY EVENT UPDATE CITIZEN QUESTIONS REGARDING RECYCLING LAMBERT BUILDING ROOF SENATE BILL 409 AND HOUSE BILL 714 (E-911 FUNDING) UPDATE TIGER FUNDING OPPORTUNITIES REVOLVING LOAN FUND LOAN TO ATSU CENVEO LIGHTING IMPROVEMENT PROJECT UPDATE BUSINESS LICENSE RENEWALS K-REDI PURCHASES LAND ADJACENT TO INDUSTRIAL PARK CONCRETE PLANTERS FOR DOWNTOWN ROTARY PARK CROSSING / BRIDGE KBSA OPENING DAY ATSU ST. LOUIS DENTAL CLINIC FISHING TOURNAMENTS ON FOREST LAKE KIRKSVILLE CELEBRATED ARBOR DAY On Friday, April 24, at 10 am, the City, Truman State University (TSU), the Missouri Department of Conservation (MDC), the Kirksville R-III School District, and the Green Thumb Project celebrated Arbor Day with a tree planting ceremony. The tree planting ceremony took place near the Outdoor Learning Center at the Kirksville R-III School with Mayor Richard Detweiler in attendance. Students from Ray Miller Elementary assisted with the planting of 2 trees and a total of 400 willow and cottonwood stakes. In honor of Arbor Day, the Green Thumb Classes at Ray Miller learned about trees for the entire week. The Arbor Day Celebration is a part of Kirksville's commitment to effective urban forest management that has helped to earn us our 17th consecutive year designation as a 2014 Tree City USA by the Arbor Day Foundation. Kirksville has met all four of the program's requirements including: a tree board or department, a tree-care ordinance, an annual community forestry budget of at least $2 per capita, and an Arbor Day observance and proclamation. -
Connect Series Accessories
Connect Series Accessories Our energy working for you. ™ power.cummins.com 2015 Cummins Power Generation Inc. All rights reserved. Specifications are subject to change without notice. Cummins Power Generation and Cummins are registered trademarks of Cummins Inc. “Our energy working for you.” is a trademark of Cummins Power Generation. Other company, product, or service names may be trademarks or service marks of others. NAAC-6260-EN (A0546T959) (01/17) CONNECT SERIES ACCESSORIES Table of Contents ALTERNATOR HEATERS ........................................................................................................ 1 ANNUNCIATORS ...................................................................................................................... 2 AUXILIARY ................................................................................................................................ 3 AUXILIARY CONFIGURABLE SIGNAL INPUTS / RELAY OUTPUTS ........................................................................ 3 AUXILIARY OUTPUTS RELAYS ....................................................................................................................... 3 BATTERIES .............................................................................................................................. 4 BATTERY ACCESSORIES ....................................................................................................... 4 BATTERY HEATER KITS ...............................................................................................................................