Implantation Period Using the Affymetrix Genechip Mirna 4.0 Array
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

A Common Analgesic Enhances the Anti-Tumour Activity of 5-Aza-2’- Deoxycytidine Through Induction of Oxidative Stress
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.31.017947; this version posted April 1, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A common analgesic enhances the anti-tumour activity of 5-aza-2’- deoxycytidine through induction of oxidative stress Hannah J. Gleneadie1,10, Amy H. Baker1, Nikolaos Batis2, Jennifer Bryant2, Yao Jiang3, Samuel J.H. Clokie4, Hisham Mehanna2, Paloma Garcia5, Deena M.A. Gendoo6, Sally Roberts5, Alfredo A. Molinolo7, J. Silvio Gutkind8, Ben A. Scheven1, Paul R. Cooper1, Farhat L. Khanim9 and Malgorzata Wiench1, 5,*. 1School of Dentistry, Institute of Clinical Studies, College of Medical and Dental Sciences, The University of Birmingham, Birmingham, B5 7EG, UK; 2Institute of Head and Neck Studies and Education (InHANSE), The University of Birmingham, Birmingham, B15 2TT, UK; 3School of Biosciences, The University of Birmingham, Birmingham, B15 2TT, UK; 4West Midlands Regional Genetics Laboratory, Birmingham Women’s and Children’s Hospital, Birmingham, B15 2TG, UK; 5Institute of Cancer and Genomic Sciences, College of Medical and Dental Sciences, The University of Birmingham, Birmingham, B15 2TT, UK; 6Centre for Computational Biology, Institute of Cancer and Genomic Sciences, The University of Birmingham, Birmingham, B15 2TT, UK; 7Moores Cancer Center and Department of Pathology, University of California San Diego, La Jolla, CA 92093, USA; 8Department of Pharmacology and Moores Cancer -

Whole Genome Sequencing of Familial Non-Medullary Thyroid Cancer Identifies Germline Alterations in MAPK/ERK and PI3K/AKT Signaling Pathways
biomolecules Article Whole Genome Sequencing of Familial Non-Medullary Thyroid Cancer Identifies Germline Alterations in MAPK/ERK and PI3K/AKT Signaling Pathways Aayushi Srivastava 1,2,3,4 , Abhishek Kumar 1,5,6 , Sara Giangiobbe 1, Elena Bonora 7, Kari Hemminki 1, Asta Försti 1,2,3 and Obul Reddy Bandapalli 1,2,3,* 1 Division of Molecular Genetic Epidemiology, German Cancer Research Center (DKFZ), D-69120 Heidelberg, Germany; [email protected] (A.S.); [email protected] (A.K.); [email protected] (S.G.); [email protected] (K.H.); [email protected] (A.F.) 2 Hopp Children’s Cancer Center (KiTZ), D-69120 Heidelberg, Germany 3 Division of Pediatric Neurooncology, German Cancer Research Center (DKFZ), German Cancer Consortium (DKTK), D-69120 Heidelberg, Germany 4 Medical Faculty, Heidelberg University, D-69120 Heidelberg, Germany 5 Institute of Bioinformatics, International Technology Park, Bangalore 560066, India 6 Manipal Academy of Higher Education (MAHE), Manipal, Karnataka 576104, India 7 S.Orsola-Malphigi Hospital, Unit of Medical Genetics, 40138 Bologna, Italy; [email protected] * Correspondence: [email protected]; Tel.: +49-6221-42-1709 Received: 29 August 2019; Accepted: 10 October 2019; Published: 13 October 2019 Abstract: Evidence of familial inheritance in non-medullary thyroid cancer (NMTC) has accumulated over the last few decades. However, known variants account for a very small percentage of the genetic burden. Here, we focused on the identification of common pathways and networks enriched in NMTC families to better understand its pathogenesis with the final aim of identifying one novel high/moderate-penetrance germline predisposition variant segregating with the disease in each studied family. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Analysis of the Impact of Pufas on Placental Gene Expression V2.28 Uploadx
TECHNISCHE UNIVERSITÄT MÜNCHEN Lehrstuhl für Ernährungsmedizin Analysis of the Impact of Polyunsaturated Fatty Acids (PUFAs) on Placental Gene Expression Eva-Maria Sedlmeier Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzender: Univ.-Prof. Dr. M. Klingenspor Prüfer der Dissertation: 1. Univ.-Prof. Dr. J. J. Hauner 2. Univ.-Prof. Dr. H. Daniel Die Dissertation wurde am 17.10.2013 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 12.12.2013 angenommen. Table of contents Table of contents Table of contents ....................................................................................................... I Summary .................................................................................................................. VI Zusammenfassung ................................................................................................ VIII 1. Introduction ........................................................................................................ 1 1.1. Fetal programming - a strategy to prevent the obesity epidemic? ...........................................1 1.1.1. The obesity epidemic ........................................................................................................1 1.1.2. Fetal programming -

S Figure 1. Cellular and Molecular Presence of B Cells, Inkt Cells, and Δγtcr T Cells in Allergic Inflammatory Tissue
Supplementary Figures: S Figure 1. Cellular and molecular presence of B cells, iNKT cells, and δγTCR T cells in allergic inflammatory tissue. A, CD45+ events isolated from biopsy tissue and autologous blood were gated and double-plotted for CD19 (for B cells) and CD3 (for T cells) in the context of normal (N) and active disease (A). B, Cellular presence of iNKT cells, identified by the invariant TCR Vα24-Jα18, as assessed in active disease biopsy and autologous blood tissue, with a spike-in of the iNKT cell line (upper panel) as the positive staining control. C, The % CD4 frequency of δγT cells was assessed by FACS (anti-δγTCR staining) in the context of normal vs. active disease. D, The frequencies of δγT cells were also assessed by extracting the entire TCR sequence pool from scRNA-seq of the 1088 tissue T cells, followed by computerized enumeration. N, normal; R, remission; A, active EoE. 1 S Figure 2. Gene ontology analysis of the tissue lymphocyte-specific genes in disease context. The full list of GO function nodes shown with two approaches, namely enrichment priority (A) and FDR- adjusted p value priority (B), with the radar scanning map representing the top contributing biofunction nodes derived from the 331 tissue-specific genes. C, The signatures of remission (R) tissue lymphocytes signature were compared to those of active (A) and normal (N) tissue lymphocytes genome wide (Mann- Whitney test, FDR-adjusted p < 0.05, fold change > 2), resulting in 217 and 333 significant genes, respectively. Heat maps in the context of all tissue and disease activities types were shown. -

WO 2014/089124 Al 12 June 20 14 ( 12.06.20 14) W P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2014/089124 Al 12 June 20 14 ( 12.06.20 14) W P O P C T (51) International Patent Classification: BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, A61K 48/00 (2006.01) A61P 37/00 (2006.01) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, A61K 39/395 (2006.01) HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2013/072938 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, 3 December 2013 (03.12.2013) TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of regional protection available): ARIPO (BW, GH, (30) Priority Data: GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, SZ, TZ, 61/732,746 3 December 2012 (03. 12.2012) US UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 61/747,653 31 December 2012 (3 1. 12.2012) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, ΓΓ, LT, LU, LV, (71) Applicants: CENEXYS, INC. -

Deep RNA Sequencing Analysis of Readthrough Gene Fusions in Human
Nacu et al. BMC Medical Genomics 2011, 4:11 http://www.biomedcentral.com/1755-8794/4/11 RESEARCHARTICLE Open Access Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples Serban Nacu, Wenlin Yuan, Zhengyan Kan, Deepali Bhatt, Celina Sanchez Rivers, Jeremy Stinson, Brock A Peters, Zora Modrusan, Kenneth Jung, Somasekar Seshagiri*, Thomas D Wu* Abstract Background: Readthrough fusions across adjacent genes in the genome, or transcription-induced chimeras (TICs), have been estimated using expressed sequence tag (EST) libraries to involve 4-6% of all genes. Deep transcriptional sequencing (RNA-Seq) now makes it possible to study the occurrence and expression levels of TICs in individual samples across the genome. Methods: We performed single-end RNA-Seq on three human prostate adenocarcinoma samples and their corresponding normal tissues, as well as brain and universal reference samples. We developed two bioinformatics methods to specifically identify TIC events: a targeted alignment method using artificial exon-exon junctions within 200,000 bp from adjacent genes, and genomic alignment allowing splicing within individual reads. We performed further experimental verification and characterization of selected TIC and fusion events using quantitative RT-PCR and comparative genomic hybridization microarrays. Results: Targeted alignment against artificial exon-exon junctions yielded 339 distinct TIC events, including 32 gene pairs with multiple isoforms. The false discovery rate was estimated to be 1.5%. Spliced alignment to the genome was less sensitive, finding only 18% of those found by targeted alignment in 33-nt reads and 59% of those in 50-nt reads. However, spliced alignment revealed 30 cases of TICs with intervening exons, in addition to distant inversions, scrambled genes, and translocations. -

Identification of Functional Genetic Variants in Inflammatory Bowel Disease by Genome and Transcriptome Sequencing
Identification of Functional Genetic Variants in Inflammatory Bowel Disease by Genome and Transcriptome Sequencing Dissertation zur Erlangung des Doktorgrades der Mathematisch-Naturwissenschaftlichen Fakultät der Christian-Albrechts-Universität zu Kiel vorgelegt von Matthias Barann August, 2012 Kiel, Deutschland 1 2 Referent/in: Prof. Dr. Philip Rosenstiel Korreferent/in: Prof. Dr. Dr. h.c. Thomas Bosch Tag der mündlichen Prüfung: 31.10.2012 Zum Druck genehmigt: 31.10.2012 gez. Prof. Dr. Wolfgang J. Duschl, Dekan 3 Parts of this dissertation are contained in the following two manuscripts. Rosenstiel R, Barann M, Klostermeier UC, Sheth V, Ellinghaus D, Rausch T, Korbel J, Nothnagel M, Krawczak M, Gilissen C, Veltman J, Forster M, Stade B, McLaughlin S, Lee CC, Fritscher-Ravens A, Franke A, Schreiber S. Whole Genome Sequence of a Crohn disease trio Barann M, Klostermeier UC, Esser D, Ammerpohl O, Siebert R, Sudbrak R, Lehrach H, Schreiber S, Rosenstiel R. Janus – Investigating the two faces of transcription 4 Contents 1. Introduction ............................................................................................................................. 7 1.1. Pathogenesis of inflammatory bowel diseases ...................................................................................... 8 1.2. Physical barrier function of the gut ......................................................................................................... 11 1.3. Pathogen recognition .................................................................................................................................. -

Cellular Features Predicting Susceptibility to Ferroptosis: Insights from Cancer Cell-Line Profiling
Cellular features predicting susceptibility to ferroptosis: insights from cancer cell-line profiling Vasanthi Sridhar Viswanathan Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2015 © 2015 Vasanthi Sridhar Viswanathan All rights reserved ABSTRACT Cellular features predicting susceptibility to ferroptosis: insights from cancer cell-line profiling Vasanthi Sridhar Viswanathan Ferroptosis is a novel non-apoptotic, oxidative form of regulated cell death that can be triggered by diverse small-molecule ferroptosis inducers (FINs) and genetic perturbations. Current lack of insights into the cellular contexts governing sensitivity to ferroptosis has hindered both translation of FINs as anti-cancer agents for specific indications and the discovery of physiological contexts where ferroptosis may function as a form of programmed cell death. This dissertation describes the identification of cellular features predicting susceptibility to ferroptosis from data generated through a large-scale profiling experiment that screened four FINs against a panel of 860 omically-characterized cancer cell lines (Cancer Therapeutics Response Portal Version 2; CTRPv2 at http://www.broadinstitute.org/ctrp/). Using correlative approaches incorporating transcriptomic, metabolomic, proteomic, and gene-dependency feature types, I uncover both pan-lineage and lineage-specific features mediating cell-line response to FINs. The first key finding from these analyses implicates high expression of sulfur and selenium metabolic pathways in conferring resistance to FINs across lineages. In contrast, the transsulfuration pathway, which enables de novo cysteine synthesis, appears to plays a role in ferroptosis resistance in a subset of lineages. The second key finding from these studies identifies cancer cells in a high mesenchymal state as being uniquely primed to undergo ferroptosis. -
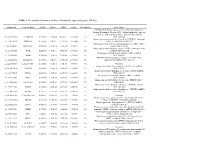
Table 1 the Statistical Metrics for Key Differentially Expressed Genes (Degs)
Table 1 The statistical metrics for key differentially expressed genes (DEGs) Agiliant Id Gene Symbol logFC pValue FDR tvalue Regulation Gene Name PREDICTED: Homo sapiens similar to Filamin-C (Gamma- filamin) (Filamin-2) (Protein FLNc) (Actin-binding-like protein) (ABP-L) (ABP-280-like protein) (LOC649056), mRNA A_24_P237896 LOC649056 0.509843 9.18E-14 4.54E-11 12.07302 Up [XR_018580] Homo sapiens programmed cell death 10 (PDCD10), transcript A_23_P18325 PDCD10 0.243111 2.8E-12 9.24E-10 10.62808 Up variant 1, mRNA [NM_007217] Homo sapiens Rho GTPase activating protein 27 (ARHGAP27), A_23_P141335 ARHGAP27 0.492709 3.97E-12 1.22E-09 10.48571 Up mRNA [NM_199282] Homo sapiens tubby homolog (mouse) (TUB), transcript variant A_23_P53110 TUB 0.528219 1.77E-11 4.56E-09 9.891033 Up 1, mRNA [NM_003320] Homo sapiens MyoD family inhibitor (MDFI), mRNA A_23_P42168 MDFI 0.314474 1.81E-10 3.74E-08 8.998697 Up [NM_005586] PREDICTED: Homo sapiens hypothetical LOC644701 A_32_P56890 LOC644701 0.444703 3.6E-10 7.09E-08 8.743973 Up (LOC644701), mRNA [XM_932316] A_32_P167111 A_32_P167111 0.873588 7.41E-10 1.4E-07 8.47781 Up Unknown Homo sapiens zinc finger protein 784 (ZNF784), mRNA A_24_P221424 ZNF784 0.686781 9.18E-10 1.68E-07 8.399687 Up [NM_203374] Homo sapiens lin-28 homolog (C. elegans) (LIN28), mRNA A_23_P74895 LIN28 0.218876 1.27E-09 2.24E-07 8.282224 Up [NM_024674] Homo sapiens ribosomal protein L5 (RPL5), mRNA A_23_P12140 RPL5 0.247598 1.81E-09 3.11E-07 8.154317 Up [NM_000969] Homo sapiens cDNA FLJ43841 fis, clone TESTI4006137.