Introducing Ribosomal Tandem Repeat Barcoding for Fungi
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Asymptomatic Infection of the Fungal Pathogen Batrachochytrium
www.nature.com/scientificreports OPEN Asymptomatic infection of the fungal pathogen Batrachochytrium salamandrivorans in captivity Received: 5 July 2017 Joana Sabino-Pinto 1, Michael Veith2, Miguel Vences 1 & Sebastian Steinfartz1 Accepted: 14 July 2018 One of the most important factors driving amphibian declines worldwide is the infectious disease, Published: xx xx xxxx chytridiomycosis. Two fungi have been associated with this disease, Batrachochytrium dendrobatidis and B. salamandrivorans (Bsal). The latter has recently driven Salamandra salamandra populations to extirpation in parts of the Netherlands, and Belgium, and potentially also in Germany. Bsal has been detected in the pet trade, which has been hypothesized to be the pathway by which it reached Europe, and which may continuously contribute to its spread. In the present study, 918 amphibians belonging to 20 captive collections in Germany and Sweden were sampled to explore the extent of Bsal presence in captivity. The fungus was detected by quantitative Polymerase Chain Reaction (qPCR) in ten collections, nine of which lacked clinical symptoms. 23 positives were confrmed by independent processing of duplicate swabs, which were analysed in a separate laboratory, and/or by sequencing ITS and 28 S gene segments. These asymptomatic positives highlight the possibility of Bsal being widespread in captive collections, and is of high conservation concern. This fnding may increase the likelihood of the pathogen being introduced from captivity into the wild, and calls for according biosecurity measures. The detection of Bsal-positive alive specimens of the hyper-susceptible fre salamander could indicate the existence of a less aggressive Bsal variant or the importance of environmental conditions for infection progression. -

Chytrid Fungi Associated with Pollen Decomposition in Crater Lake, Oregon Kathleen A
APPLIED & ENVIRONMENTAL MICROBIOLOGY • 83 CHYTRID FUNGI ASSOCIATED WITH POLLEN DECOMPOSITION IN CRATER LAKE, OREGON KATHLEEN A. PAGE* AND MEGHAN K. FLANNERY DEPARTMENT OF BIOLOGY, SOUTHERN OREGON UNIVERSITY, ASHLAND, OR USA MANUSCRIPT RECEIVED 25 OCTOBER 2017; ACCEPTED 27 JANUARY 2018 Copyright 2018, Fine Focus. All Rights Reserved. 84 • FINE FOCUS, VOL. 4(1) ABSTRACT We identified chytrid fungi that were attached to pine pollen on the surface of Crater Lake. Fungi were identified by large subunit (LSU) rRNA gene sequencing of lake pollen extracts and by isolation of a chytrid fungus that was present on the pollen. LSU rRNA PCR products were cloned, sequenced and identified. The majority of eukaryotic LSU rRNA sequences associated with pollen were found to be members of the chytrid order Rhizophyidiales. A fungal CORRESPONDING isolate was characterized culturally, morphologically, and AUTHOR by DNA sequencing and was identified as a member of the genus Paranamyces, in the order Rhizophydiales. In addition, Kathleen A. Page protist LSU rRNA sequences from the phylum Ciliophora [email protected] were found. The concentrations of dissolved organic matter, nitrogen, and phosphate in surface water that had visible KEYWORDS pollen rafts increased according to the concentration of pollen in the water. Each of these nutrients was detected • Chytrid at several fold higher levels in water with pollen rafts as • Pollen compared to surface water lacking pollen rafts. These results • Crater Lake ecosystem • Food Webs provide evidence for the role of chytrid fungi in nutrient • Fungal Aquatic Ecology release from pollen deposited on Crater Lake. INTRODUCTION The occurrence of pollen in Crater Lake: depth of 594 m. -

Chytridiomycetes, Chytridiomycota)
VOLUME 5 JUNE 2020 Fungal Systematics and Evolution PAGES 17–38 doi.org/10.3114/fuse.2020.05.02 Taxonomic revision of the genus Zygorhizidium: Zygorhizidiales and Zygophlyctidales ord. nov. (Chytridiomycetes, Chytridiomycota) K. Seto1,2,3*, S. Van den Wyngaert4, Y. Degawa1, M. Kagami2,3 1Sugadaira Research Station, Mountain Science Center, University of Tsukuba, 1278-294, Sugadaira-Kogen, Ueda, Nagano 386-2204, Japan 2Department of Environmental Science, Faculty of Science, Toho University, 2-2-1, Miyama, Funabashi, Chiba 274-8510, Japan 3Graduate School of Environment and Information Sciences, Yokohama National University, 79-7, Tokiwadai, Hodogaya, Yokohama, Kanagawa 240- 8502, Japan 4Department of Experimental Limnology, Leibniz-Institute of Freshwater Ecology and Inland Fisheries, Alte Fischerhuette 2, D-16775 Stechlin, Germany *Corresponding author: [email protected] Key words: Abstract: During the last decade, the classification system of chytrids has dramatically changed based on zoospore Chytridiomycota ultrastructure and molecular phylogeny. In contrast to well-studied saprotrophic chytrids, most parasitic chytrids parasite have thus far been only morphologically described by light microscopy, hence they hold great potential for filling taxonomy some of the existing gaps in the current classification of chytrids. The genus Zygorhizidium is characterized by an zoospore ultrastructure operculate zoosporangium and a resting spore formed as a result of sexual reproduction in which a male thallus Zygophlyctis and female thallus fuse via a conjugation tube. All described species of Zygorhizidium are parasites of algae and Zygorhizidium their taxonomic positions remain to be resolved. Here, we examined morphology, zoospore ultrastructure, host specificity, and molecular phylogeny of seven cultures of Zygorhizidium spp. Based on thallus morphology and host specificity, one culture was identified as Z. -

Introducing Ribosomal Tandem Repeat Barcoding for Fungi
bioRxiv preprint doi: https://doi.org/10.1101/310540; this version posted April 28, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Introducing ribosomal tandem repeat barcoding for fungi Christian Wurzbacher1,2,3, Ellen Larsson1,3, Johan Bengtsson-Palme4,5, Silke Van den Wyngaert6, Sten Svantesson1,3, Erik Kristiansson7, Maiko Kagami6,8,9, R. Henrik Nilsson1,3 Affiliations 1. Department of Biological and Environmental Sciences, University of Gothenburg, Box 461, 40530 Göteborg, Sweden. 2. Chair of Urban Water Systems Engineering, Technical University of Munich, Am Coulombwall 3, Garching 85748, Germany 3. Gothenburg Global Biodiversity Centre, Box 461, 405 30 Göteborg, Sweden 4. Wisconsin Institute for Discovery, University of Wisconsin-Madison, 330 North Orchard Street, Madison WI 53715, Wisconsin, USA. 5. Department of Infectious Diseases, Institute of Biomedicine, The Sahlgrenska Academy, University of Gothenburg, Guldhedsgatan 10, 413 46, Göteborg, Sweden 6. Leibniz-Institute of Freshwater Ecology and Inland Fisheries Berlin, Alte Fischerhütte 2, 16775 Stechlin, Germany 7. Department of Mathematical Sciences, Chalmers University of Technology and University of Gothenburg, 412 96 Göteborg, Sweden 8. Department of Environmental Science, Faculty of Science, Toho University, 2-2-1 Miyama, Funabashi, Chiba, Japan 9. Graduate School of Environmental and Information Sciences, Yokohama National University, Tokiwadai 79- 7, Hodogayaku, Yokohama, Kanagawa, Japan Abstract Sequence analysis of the various ribosomal genetic markers is the dominant molecular method for identification and description of fungi. -

Zoosporic Parasites Infecting Marine Diatoms E a Black Box That Needs to Be Opened
fungal ecology xxx (2015) 1e18 available at www.sciencedirect.com ScienceDirect journal homepage: www.elsevier.com/locate/funeco Zoosporic parasites infecting marine diatoms e A black box that needs to be opened Bettina SCHOLZa,b, Laure GUILLOUc, Agostina V. MARANOd, Sigrid NEUHAUSERe, Brooke K. SULLIVANf, Ulf KARSTENg, € h i, Frithjof C. KUPPER , Frank H. GLEASON * aBioPol ehf., Einbuastig 2, 545 Skagastrond,€ Iceland bFaculty of Natural Resource Sciences, University of Akureyri, Borgir v. Nordurslod, IS 600 Akureyri, Iceland cSorbonne Universites, Universite Pierre et Marie Curie e Paris 6, UMR 7144, Station Biologique de Roscoff, Place Georges Teissier, CS90074, 29688 Roscoff cedex, France dInstituto de Botanica,^ Nucleo de Pesquisa em Micologia, Av. Miguel Stefano 3687, 04301-912, Sao~ Paulo, SP, Brazil eInstitute of Microbiology, University of Innsbruck, Technikerstr. 25, A-6020 Innsbruck, Austria fDepartment of Biosciences, University of Melbourne, Parkville, VIC 3010, Australia gInstitute of Biological Sciences, Applied Ecology & Phycology, University of Rostock, Albert-Einstein-Strasse 3, 18059 Rostock, Germany hOceanlab, University of Aberdeen, Main Street, Newburgh AB41 6AA, Scotland, United Kingdom iSchool of Biological Sciences FO7, University of Sydney, Sydney, NSW 2006, Australia article info abstract Article history: Living organisms in aquatic ecosystems are almost constantly confronted by pathogens. Received 12 May 2015 Nevertheless, very little is known about diseases of marine diatoms, the main primary Revision received 2 September 2015 producers of the oceans. Only a few examples of marine diatoms infected by zoosporic Accepted 2 September 2015 parasites are published, yet these studies suggest that diseases may have significant Available online - impacts on the ecology of individual diatom hosts and the composition of communities at Corresponding editor: both the producer and consumer trophic levels of food webs. -

Early Diverging Lineages Within Cryptomycota and Chytridiomycota Dominate the Fungal Communities in Ice-Covered Lakes of the Mcmurdo Dry Valleys, Antarctica
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/320986652 Early diverging lineages within Cryptomycota and Chytridiomycota dominate the fungal communities in ice-covered lakes of the McMurdo Dry Valleys, Antarctica Article in Scientific Reports · November 2017 DOI: 10.1038/s41598-017-15598-w CITATIONS READS 2 144 6 authors, including: Keilor Rojas- Jimenez Christian Wurzbacher University of Costa Rica Technische Universität München 28 PUBLICATIONS 289 CITATIONS 59 PUBLICATIONS 398 CITATIONS SEE PROFILE SEE PROFILE Elizabeth Bourne Amy Chiuchiolo Leibniz-Institute of Freshwater Ecology and Inland Fisheries Montana State University 9 PUBLICATIONS 450 CITATIONS 11 PUBLICATIONS 322 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: MANTEL View project HGT in aquatic ecosystems View project All content following this page was uploaded by Keilor Rojas-Jimenez on 10 November 2017. The user has requested enhancement of the downloaded file. www.nature.com/scientificreports OPEN Early diverging lineages within Cryptomycota and Chytridiomycota dominate the fungal communities Received: 25 August 2017 Accepted: 30 October 2017 in ice-covered lakes of the McMurdo Published: xx xx xxxx Dry Valleys, Antarctica Keilor Rojas-Jimenez 1,2, Christian Wurzbacher1,3, Elizabeth Charlotte Bourne3,4, Amy Chiuchiolo5, John C. Priscu5 & Hans-Peter Grossart 1,6 Antarctic ice-covered lakes are exceptional sites for studying the ecology of aquatic fungi under conditions of minimal human disturbance. In this study, we explored the diversity and community composition of fungi in fve permanently covered lake basins located in the Taylor and Miers Valleys of Antarctica. -

Lepelletier Et Al 2014 Dinomyces Arenysensis Gen.Pdf
Protist, Vol. 165, 230–244, March 2014 http://www.elsevier.de/protis Published online date 1 March 2014 ORIGINAL PAPER Dinomyces arenysensis gen. et sp. nov. (Rhizophydiales, Dinomycetaceae fam. nov.), a Chytrid Infecting Marine Dinoflagellates a,b c,d e f,b Frédéric Lepelletier , Sergey A. Karpov , Elisabet Alacid , Sophie Le Panse , a,b e a,b a,b,1 Estelle Bigeard , Esther Garcés , Christian Jeanthon , and Laure Guillou a CNRS, UMR 7144, Place Georges Teissier, CS90074, 29688 Roscoff Cedex, France b Université Pierre et Marie Curie (Paris VI), Station Biologique de Roscoff, Place Georges Teissier, CS90074, 29688 Roscoff Cedex, France c Zoological Institute RAS, St. Petersburg, Russia d St. Petersburg State University, St. Petersburg, Russia e Departament de Biologia Marina i Oceanografia, Institut de Ciències del Mar, CSIC, Pg. Marítim de la Barceloneta, 37-49, E08003 Barcelona, Spain f CNRS, FR 2424, Plate-forme Merimage, Station Biologique de Roscoff, Place Georges Teissier, CS90074, 29688 Roscoff Cedex, France Submitted January 14, 2014; Accepted February 21, 2014 Monitoring Editor: Michael Melkonian Environmental 18S rRNA gene surveys of microbial eukaryotes have recently revealed the diversity of major parasitic agents in pelagic freshwater systems, consisting primarily of chytrid fungi. To date, only a few studies have reported the presence of chydrids in the marine environment and a limited number of marine chytrids have been properly identified and characterized. Here, we report the isolation and cul- tivation of a marine chytrid from samples taken during a bloom of the toxic dinoflagellate Alexandrium minutum in the Arenys de Mar harbour (Mediterranean Sea, Spain). Cross-infections using cultures and natural phytoplankton communities revealed that this chytrid is only able to infect certain species of dinoflagellates, with a rather wide host range but with a relative preference for Alexandrium species. -

Ultrastructural and Molecular Analyses of Rhizophydiales (Chytridiomycota) Isolates from North America and Argentina
mycological research 112 (2008) 759–782 journal homepage: www.elsevier.com/locate/mycres Ultrastructural and molecular analyses of Rhizophydiales (Chytridiomycota) isolates from North America and Argentina Peter M. LETCHERa,*, Carlos G. VE´LEZb, Marı´a Eugenia BARRANTESb, Martha J. POWELLa, Perry F. CHURCHILLa, William S. WAKEFIELDa aDepartment of Biological Sciences, The University of Alabama, Tuscaloosa, AL 35487, USA bDepartamento de Biodiversidad y Biologia Experimental, Universidad de Buenos Aires, C1428EHA Buenos Aires, Argentina article info abstract Article history: The Rhizophydiales is the most recently circumscribed order in the Chytridiomycota. Past Received 12 July 2007 studies focused on soil chytrids from North America and Australia to determine the range Received in revised form of diversity within this clade of chytrids and established three families (Rhizophydiaceae, 9 November 2007 Terramycetaceae, and Kappamycetaceae) in the new order. Although Rhizophydiales contains Accepted 24 January 2008 seemingly simple chytrids morphologically, analyses of ribosomal gene sequences and Corresponding Editor: zoospore characters have demonstrated unexpected genetic and ultrastructural diversity, David L. Hawksworth highlighting the need for broader habitat and geographic sampling to reveal the actual diversity within this new order. To enlarge our sampling, in this study we investigated Keywords: 38 newly cultured chytrids collected from aquatic habitats in Argentina, a territory Chytrid under-explored for chytrid diversity. From analyses of thallus morphology, zoospore ultra- Phylogeny structure, and 28S and ITS1–5.8S–ITS2 ribosomal gene sequences, we expand the concept Ribosomal genes of Rhizophydiales, describing seven new families (Alphamycetaceae, Angulomycetaceae, Aqua- Taxonomy mycetaceae, Globomycetaceae, Gorgonomycetaceae, Pateramycetaceae, and Protrudomycetaceae) Zoospore and eight new genera (Alphamyces, Angulomyces, Aquamyces, Globomyces, Urceomyces, Gorgo- nomyces, Pateramyces, and Protrudomyces). -

Toward a Fully Resolved Fungal Tree of Life
Annual Review of Microbiology Toward a Fully Resolved Fungal Tree of Life Timothy Y. James,1 Jason E. Stajich,2 Chris Todd Hittinger,3 and Antonis Rokas4 1Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, Michigan 48109, USA; email: [email protected] 2Department of Microbiology and Plant Pathology, Institute for Integrative Genome Biology, University of California, Riverside, California 92521, USA; email: [email protected] 3Laboratory of Genetics, DOE Great Lakes Bioenergy Research Center, Wisconsin Energy Institute, Center for Genomic Science and Innovation, J.F. Crow Institute for the Study of Evolution, University of Wisconsin–Madison, Madison, Wisconsin 53726, USA; email: [email protected] 4Department of Biological Sciences, Vanderbilt University, Nashville, Tennessee 37235, USA; email: [email protected] Annu. Rev. Microbiol. 2020. 74:291–313 Keywords First published as a Review in Advance on deep phylogeny, phylogenomic inference, uncultured majority, July 13, 2020 classification, systematics The Annual Review of Microbiology is online at micro.annualreviews.org Abstract https://doi.org/10.1146/annurev-micro-022020- Access provided by Vanderbilt University on 06/28/21. For personal use only. In this review, we discuss the current status and future challenges for fully 051835 Annu. Rev. Microbiol. 2020.74:291-313. Downloaded from www.annualreviews.org elucidating the fungal tree of life. In the last 15 years, advances in genomic Copyright © 2020 by Annual Reviews. technologies have revolutionized fungal systematics, ushering the field into All rights reserved the phylogenomic era. This has made the unthinkable possible, namely ac- cess to the entire genetic record of all known extant taxa. -

Article.Pdf (4.860Mb)
Mycologia ISSN: (Print) (Online) Journal homepage: https://www.tandfonline.com/loi/umyc20 New species of aquatic chytrids from Oman Brandon T. Hassett , Badriya K. Al-Shaibi , Abdulrahman Al-Nabhani & Abdullah M. Al-Sadi To cite this article: Brandon T. Hassett , Badriya K. Al-Shaibi , Abdulrahman Al-Nabhani & Abdullah M. Al-Sadi (2020) New species of aquatic chytrids from Oman, Mycologia, 112:4, 781-791, DOI: 10.1080/00275514.2020.1761226 To link to this article: https://doi.org/10.1080/00275514.2020.1761226 © 2020 The Author(s). Published with license by Taylor & Francis Group, LLC. View supplementary material Published online: 12 Jun 2020. Submit your article to this journal Article views: 195 View related articles View Crossmark data Full Terms & Conditions of access and use can be found at https://www.tandfonline.com/action/journalInformation?journalCode=umyc20 MYCOLOGIA 2020, VOL. 112, NO. 4, 781–791 https://doi.org/10.1080/00275514.2020.1761226 New species of aquatic chytrids from Oman Brandon T. Hassett a, Badriya K. Al-Shaibib, Abdulrahman Al-Nabhanib, and Abdullah M. Al-Sadi b aUiT–The Arctic University of Norway, Framstredet 6, Tromsø 9019, Norway; bDepartment of Crop Sciences, College of Agricultural and Marine Sciences, Sultan Qaboos University, P.O. Box-34, Al-Khod 123, Oman ABSTRACT ARTICLE HISTORY Oman is a desert country in the south of the Middle East. Springs and other water sources that Received 29 January 2020 harbor aquatic organisms can be separated by hundreds of kilometers. In Oct 2019, we isolated four Accepted 23 April 2020 freshwater aquatic fungi (Chytridiomycota) from benthic detritus baited with pine pollen on KEYWORDS a general nutrient medium near Salalah, Oman. -
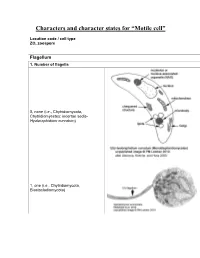
Characters and Character States for “Motile Cell”
Characters and character states for “Motile cell” Location code / cell type ZO, zoospore Flagellum 1. Number of flagella 0, none (i.e., Chytridiomycota, Chytridiomycetes: incertae sedis- Hyaloraphidium curvatum) 1, one (i.e., Chytridiomycota, Blastocladiomycota) 2, multiple (i.e., Neocallimastigomycota) 2. Electron-opaque plug in axoneme core and between axoneme and flagellar membrane 0, absent 1, present (i.e., Chytridiomycota: Chytridiomycetes: Chytridiales, Lobulomycetales, Cladochytriales, incertae sedis: Synchytrium endobioticum, Polychytrium clade; Monoblepharidomycetes) 3. Flagellum coating 0, absent (all taxa except Polyphagus euglenae) 1, present (Polyphagus euglenae) Kinetosome (the term “basal body” is synonymous; Andersen et al. 1991) 4. Electron-opaque core in kinetosome 0, absent (all taxa except Kappamyces) 1, present (Chytridiomycota: Chytridiomycetes: Rhizophydiales- Kappamyces) 5. Scalloped ring within kinetosome, extensions of the A, B, or C microtubule 0, absent 1, present (Lacustromyces hiemalis) Kinetosome-associated structures 6. Kinetosome support 0, absent (Thalassochytrium gracilaripsidis) 1, kinetosome props 2, broken kinetosome props (Olpidium radicale) 3, skirt-like structure surrounding kinetosome (Neocallimastigomycota) 7. Kinetosome-associated plates 0, absent 1, present (Chytridiomycota: Chytridiomycetes: Chytridiales- Group I- type zoospore [Barr 1980]) 8. Kinetosome-associated saddle 0, absent 1, present (Chytridiomycota: Chytridiomycetes: Chytridiales- Group II-type zoospore [Barr 1980]- Chytridium -

Taxonomic Revision of the Genus <I> Zygorhizidium</I>: <I> Zygorhizidiales</I> and <I> Zygophlycti
VOLUME 5 JUNE 2020 Fungal Systematics and Evolution PAGES 17–38 doi.org/10.3114/fuse.2020.05.02 Taxonomic revision of the genus Zygorhizidium: Zygorhizidiales and Zygophlyctidales ord. nov. (Chytridiomycetes, Chytridiomycota) K. Seto1,2,3*, S. Van den Wyngaert4, Y. Degawa1, M. Kagami2,3 1Sugadaira Research Station, Mountain Science Center, University of Tsukuba, 1278-294, Sugadaira-Kogen, Ueda, Nagano 386-2204, Japan 2Department of Environmental Science, Faculty of Science, Toho University, 2-2-1, Miyama, Funabashi, Chiba 274-8510, Japan 3Graduate School of Environment and Information Sciences, Yokohama National University, 79-7, Tokiwadai, Hodogaya, Yokohama, Kanagawa 240- 8502, Japan 4Department of Experimental Limnology, Leibniz-Institute of Freshwater Ecology and Inland Fisheries, Alte Fischerhuette 2, D-16775 Stechlin, Germany *Corresponding author: [email protected] Key words: Abstract: During the last decade, the classification system of chytrids has dramatically changed based on zoospore Chytridiomycota ultrastructure and molecular phylogeny. In contrast to well-studied saprotrophic chytrids, most parasitic chytrids parasite have thus far been only morphologically described by light microscopy, hence they hold great potential for filling taxonomy some of the existing gaps in the current classification of chytrids. The genus Zygorhizidium is characterized by an zoospore ultrastructure operculate zoosporangium and a resting spore formed as a result of sexual reproduction in which a male thallus Zygophlyctis and female thallus fuse via a conjugation tube. All described species of Zygorhizidium are parasites of algae and Zygorhizidium their taxonomic positions remain to be resolved. Here, we examined morphology, zoospore ultrastructure, host specificity, and molecular phylogeny of seven cultures of Zygorhizidium spp. Based on thallus morphology and host specificity, one culture was identified as Z.