Recent Progress in Reconstructing Lophotrochozoan (Spiralian) Phylogeny
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Systema Naturae∗
Systema Naturae∗ c Alexey B. Shipunov v. 5.802 (June 29, 2008) 7 Regnum Monera [ Bacillus ] /Bacteria Subregnum Bacteria [ 6:8Bacillus ]1 Superphylum Posibacteria [ 6:2Bacillus ] stat.m. Phylum 1. Firmicutes [ 6Bacillus ]2 Classis 1(1). Thermotogae [ 5Thermotoga ] i.s. 2(2). Mollicutes [ 5Mycoplasma ] 3(3). Clostridia [ 5Clostridium ]3 4(4). Bacilli [ 5Bacillus ] 5(5). Symbiobacteres [ 5Symbiobacterium ] Phylum 2. Actinobacteria [ 6Actynomyces ] Classis 1(6). Actinobacteres [ 5Actinomyces ] Phylum 3. Hadobacteria [ 6Deinococcus ] sed.m. Classis 1(7). Hadobacteres [ 5Deinococcus ]4 Superphylum Negibacteria [ 6:2Rhodospirillum ] stat.m. Phylum 4. Chlorobacteria [ 6Chloroflexus ]5 Classis 1(8). Ktedonobacteres [ 5Ktedonobacter ] sed.m. 2(9). Thermomicrobia [ 5Thermomicrobium ] 3(10). Chloroflexi [ 5Chloroflexus ] ∗Only recent taxa. Viruses are not included. Abbreviations and signs: sed.m. (sedis mutabilis); stat.m. (status mutabilis): s., aut i. (superior, aut interior); i.s. (incertae sedis); sed.p. (sedis possibilis); s.str. (sensu stricto); s.l. (sensu lato); incl. (inclusum); excl. (exclusum); \quotes" for environmental groups; * (asterisk) for paraphyletic taxa; / (slash) at margins for major clades (\domains"). 1Incl. \Nanobacteria" i.s. et dubitativa, \OP11 group" i.s. 2Incl. \TM7" i.s., \OP9", \OP10". 3Incl. Dictyoglomi sed.m., Fusobacteria, Thermolithobacteria. 4= Deinococcus{Thermus. 5Incl. Thermobaculum i.s. 1 4(11). Dehalococcoidetes [ 5Dehalococcoides ] 5(12). Anaerolineae [ 5Anaerolinea ]6 Phylum 5. Cyanobacteria [ 6Nostoc ] Classis 1(13). Gloeobacteres [ 5Gloeobacter ] 2(14). Chroobacteres [ 5Chroococcus ]7 3(15). Hormogoneae [ 5Nostoc ] Phylum 6. Bacteroidobacteria [ 6Bacteroides ]8 Classis 1(16). Fibrobacteres [ 5Fibrobacter ] 2(17). Chlorobi [ 5Chlorobium ] 3(18). Salinibacteres [ 5Salinibacter ] 4(19). Bacteroidetes [ 5Bacteroides ]9 Phylum 7. Spirobacteria [ 6Spirochaeta ] Classis 1(20). Spirochaetes [ 5Spirochaeta ] s.l.10 Phylum 8. Planctobacteria [ 6Planctomyces ]11 Classis 1(21). -

Platyhelminthes, Nemertea, and "Aschelminthes" - A
BIOLOGICAL SCIENCE FUNDAMENTALS AND SYSTEMATICS – Vol. III - Platyhelminthes, Nemertea, and "Aschelminthes" - A. Schmidt-Rhaesa PLATYHELMINTHES, NEMERTEA, AND “ASCHELMINTHES” A. Schmidt-Rhaesa University of Bielefeld, Germany Keywords: Platyhelminthes, Nemertea, Gnathifera, Gnathostomulida, Micrognathozoa, Rotifera, Acanthocephala, Cycliophora, Nemathelminthes, Gastrotricha, Nematoda, Nematomorpha, Priapulida, Kinorhyncha, Loricifera Contents 1. Introduction 2. General Morphology 3. Platyhelminthes, the Flatworms 4. Nemertea (Nemertini), the Ribbon Worms 5. “Aschelminthes” 5.1. Gnathifera 5.1.1. Gnathostomulida 5.1.2. Micrognathozoa (Limnognathia maerski) 5.1.3. Rotifera 5.1.4. Acanthocephala 5.1.5. Cycliophora (Symbion pandora) 5.2. Nemathelminthes 5.2.1. Gastrotricha 5.2.2. Nematoda, the Roundworms 5.2.3. Nematomorpha, the Horsehair Worms 5.2.4. Priapulida 5.2.5. Kinorhyncha 5.2.6. Loricifera Acknowledgements Glossary Bibliography Biographical Sketch Summary UNESCO – EOLSS This chapter provides information on several basal bilaterian groups: flatworms, nemerteans, Gnathifera,SAMPLE and Nemathelminthes. CHAPTERS These include species-rich taxa such as Nematoda and Platyhelminthes, and as taxa with few or even only one species, such as Micrognathozoa (Limnognathia maerski) and Cycliophora (Symbion pandora). All Acanthocephala and subgroups of Platyhelminthes and Nematoda, are parasites that often exhibit complex life cycles. Most of the taxa described are marine, but some have also invaded freshwater or the terrestrial environment. “Aschelminthes” are not a natural group, instead, two taxa have been recognized that were earlier summarized under this name. Gnathifera include taxa with a conspicuous jaw apparatus such as Gnathostomulida, Micrognathozoa, and Rotifera. Although they do not possess a jaw apparatus, Acanthocephala also belong to Gnathifera due to their epidermal structure. ©Encyclopedia of Life Support Systems (EOLSS) BIOLOGICAL SCIENCE FUNDAMENTALS AND SYSTEMATICS – Vol. -

Defining Phyla: Evolutionary Pathways to Metazoan Body Plans
EVOLUTION & DEVELOPMENT 3:6, 432-442 (2001) Defining phyla: evolutionary pathways to metazoan body plans Allen G. Collins^ and James W. Valentine* Museum of Paleontology and Department of Integrative Biology, University of California, Berkeley, CA 94720, USA 'Author for correspondence (email: [email protected]) 'Present address: Section of Ecology, Befiavior, and Evolution, Division of Biology, University of California, San Diego, La Jolla, CA 92093-0116, USA SUMMARY Phyla are defined by two sets of criteria, one pothesis of Nielsen; the clonal hypothesis of Dewel; the set- morphological and the other historical. Molecular evidence aside cell hypothesis of Davidson et al.; and a benthic hy- permits the grouping of animals into clades and suggests that pothesis suggested by the fossil record. It is concluded that a some groups widely recognized as phyla are paraphyletic, benthic radiation of animals could have supplied the ances- while some may be polyphyletic; the phyletic status of crown tral lineages of all but a few phyla, is consistent with molecu- phyla is tabulated. Four recent evolutionary scenarios for the lar evidence, accords well with fossil evidence, and accounts origins of metazoan phyla and of supraphyletic clades are as- for some of the difficulties in phylogenetic analyses of phyla sessed in the light of a molecular phylogeny: the trochaea hy- based on morphological criteria. INTRODUCTION Molecules have provided an important operational ad- vance to addressing questions about the origins of animal Concepts of animal phyla have changed importantly from phyla. Molecular developmental and comparative genomic their origins in the six Linnaean classis and four Cuvieran evidence offer insights into the genetic bases of body plan embranchements. -

Invertebrates Invertebrates: • Are Animals Without Backbones • Represent 95% of the Animal Kingdom Animal Diversity Morphological Vs
Invertebrates Invertebrates: • Are animals without backbones • Represent 95% of the animal kingdom Animal Diversity Morphological vs. Molecular Character Phylogeny? A tree is a hypothesis supported or not supported by evidence. Groupings change as new evidence become available. Sponges - Porifera Natural Bath Sponges – over-collected, now uncommon Sponges • Perhaps oldest animal phylum (Ctenphora possibly older) • may represent several old phyla, some now extinct ----------------Ctenophora? Sponges - Porifera • Mostly marine • Sessile animals • Lack true tissues; • Have only a few cell types, cells kind of independent • Most have no symmetry • Body resembles a sac perforated with holes, system of canals. • Strengthened by fibers of spongin, spicules Sponges have a variety of shapes Sponges Pores Choanocyte Amoebocyte (feeding cell) Skeletal Water fiber flow Central cavity Flagella Choanocyte in contact with an amoebocyte Sponges - Porifera • Sessile filter feeder • No mouth • Sac-like body, perforated by pores. • Interior lined by flagellated cells (choanocytes). Flagellated collar cells generate a current, draw water through the walls of the sponge where food is collected. • Amoeboid cells move around in the mesophyll and distribute food. Sponges - Porifera Grantia x.s. Sponge Reproduction Asexual reproduction • Fragmentation or by budding. • Sponges are capable of regeneration, growth of a whole from a small part. Sexual reproduction • Hermaphrodites, produce both eggs and sperm • Eggs and sperm released into the central cavity • Produces -

"Phoronida". In: Encyclopedia of Life Science
Phoronida Introductory article Christian C Emig, Centre d’Oce´anologie de Marseille CNRS, Marseille, France Article Contents . Basic Design The Phoronida, divided into two genera and 10 species, is a small marine group, which . Diversity and Lifestyles belongs to the phylum Lophophorata. Fossil History and Phylogeny Basic Design complexity of the lophophore from an oval to a helicoidal, The Phoronida is an exclusively marine group with a sessile through a horseshoe and spiral shape. This is related to an vermiform body enclosed in a tube (Figure 1) The body is increase in the number of tentacles, which is proportional composed of three distinct parts (prosome, mesosome and to the general body size. metasome), each containing its own coelomic cavity. The The metasome (or trunk) is slender and cylindrical, with prosome forms the epistome, a fold overhanging the mouth a bulb-like posterior end (or ampulla) that anchors the dorsally. The mesosome bears the lophophore, with the body in the rear end of the tube. It is separated from the rest mouth lying between its two rows of tentacles. The of the body by the diaphragm, a thick transverse septum lophophore is a terminal, bilaterally symmetrical, tentacle located behind the lophophore. crown, each tentacle having complex arrays of cilia for The digestive tract is U-shaped, bringing the anus close filter-feeding. Lophophore shape is a fairly constant to the mouth. The descending branch is divided into a short feature within each species, and there is an increasing oesophagus, followed by a long prestomach, then a stomach surrounded by a blood plexus. -
S I Section 4
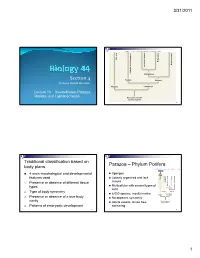
3/31/2011 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Porifera Ecdysozoa Deuterostomia Lophotrochozoa Cnidaria and Ctenophora Cnidaria and Protostomia SSiection 4 Radiata Bilateria Professor Donald McFarlane Parazoa Eumetazoa Lecture 13 Invertebrates: Parazoa, Radiata, and Lophotrochozoa Ancestral colonial choanoflagellate 2 Traditional classification based on Parazoa – Phylum Porifera body plans Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Parazoa 4 main morphological and developmental Sponges features used Loosely organized and lack Porifera Ecdysozoa Cnidaria and Ctenophora tissues euterostomia photrochozoa D 1. o Presence or absence of different tissue L Multicellular with several types of types Protostomia cells 2. Type of body symmetry Radiata Bilateria 8,000 species, mostly marine Parazoa Eumetazoa 3. Presence or absence of a true body No apparent symmetry cavity Ancestral colonial Adults sessile, larvae free- choanoflagellate 4. Patterns of embryonic development swimming 3 4 1 3/31/2011 Water drawn through pores (ostia) into spongocoel Flows out through osculum Reproduce Choanocytes line spongocoel Sexually Most hermapppgggphrodites producing eggs and sperm Trap and eat small particles and plankton Gametes are derived from amoebocytes or Mesohyl between choanocytes and choanocytes epithelial cells Asexually Amoebocytes absorb food from choanocytes, Small fragment or bud may detach and form a new digest it, and carry -

The Genome of the Poecilogonous Annelid Streblospio Benedicti Christina Zakas1, Nathan D
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.15.440069; this version posted April 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The genome of the poecilogonous annelid Streblospio benedicti Christina Zakas1, Nathan D. Harry1, Elizabeth H. Scholl2 and Matthew V. Rockman3 1Department of Genetics, North Carolina State University, Raleigh, NC, USA 2Bioinformatics Research Center, North Carolina State University, Raleigh, NC, USA 3Department of Biology and Center for Genomics & Systems Biology, New York University, New York, NY, USA [email protected] [email protected] Abstract Streblospio benedicti is a common marine annelid that has become an important model for developmental evolution. It is the only known example of poecilogony, where two distinct developmental modes occur within a single species, that is due to a heritable difference in egg size. The dimorphic developmental programs and life-histories exhibited in this species depend on differences within the genome, making it an optimal model for understanding the genomic basis of developmental divergence. Studies using S. benedicti have begun to uncover the genetic and genomic principles that underlie developmental uncoupling, but until now they have been limited by the lack of availability of genomic tools. Here we present an annotated chromosomal-level genome assembly of S. benedicti generated from a combination of Illumina reads, Nanopore long reads, Chicago and Hi-C chromatin interaction sequencing, and a genetic map from experimental crosses. At 701.4 Mb, the S. benedicti genome is the largest annelid genome to date that has been assembled to chromosomal scaffolds, yet it does not show evidence of extensive gene family expansion, but rather longer intergenic regions. -

Phylogenomics of Tubeworms (Siboglinidae, Annelida) and Comparative Performance of Different Reconstruction Methods
Zoologica Scripta Phylogenomics of tubeworms (Siboglinidae, Annelida) and comparative performance of different reconstruction methods YUANNING LI,KEVIN M. KOCOT,NATHAN V. WHELAN,SCOTT R. SANTOS,DAMIEN S. WAITS, DANIEL J. THORNHILL &KENNETH M. HALANYCH Submitted: 28 January 2016 Li, Y., Kocot, K.M., Whelan, N.V., Santos, S.R., Waits, D.S., Thornhill, D.J. & Halanych, Accepted: 18 June 2016 K.M. (2016). Phylogenomics of tubeworms (Siboglinidae, Annelida) and comparative perfor- doi:10.1111/zsc.12201 mance of different reconstruction methods. —Zoologica Scripta, 00: 000–000. Deep-sea tubeworms (Annelida, Siboglinidae) represent dominant species in deep-sea chemosynthetic communities (e.g. hydrothermal vents and cold methane seeps) and occur in muddy sediments and organic falls. Siboglinids lack a functional digestive tract as adults, and they rely on endosymbiotic bacteria for energy, making them of evolutionary and physi- ological interest. Despite their importance, inferred evolutionary history of this group has been inconsistent among studies based on different molecular markers. In particular, place- ment of bone-eating Osedax worms has been unclear in part because of their distinctive biol- ogy, including harbouring heterotrophic bacteria as endosymbionts, displaying extreme sexual dimorphism and exhibiting a distinct body plan. Here, we reconstructed siboglinid evolutionary history using 12 newly sequenced transcriptomes. We parsed data into three data sets that accommodated varying levels of missing data, and we evaluate effects of miss- ing data on phylogenomic inference. Additionally, several multispecies-coalescent approaches and Bayesian concordance analysis (BCA) were employed to allow for a compar- ison of results to a supermatrix approach. Every analysis conducted herein strongly sup- ported Osedax being most closely related to the Vestimentifera and Sclerolinum clade, rather than Frenulata, as previously reported. -

Animal Diversity Part 2
Textbook resources • pp. 517-522 • pp. 527-8 Animal Diversity • p. 530 part 2 • pp. 531-2 Clicker question In protostomes A. The blastopore becomes the mouth. B. The blastopore becomes the anus. C. Development involves indeterminate cleavage. D. B and C Fig. 25.2 Phylogeny to know (1). Symmetry Critical innovations to insert: Oral bilateral symmetry ecdysis mouth develops after anus multicellularity Aboral tissues 1 Animal diversity, part 2 Parazoa Diversity 2 I. Parazoa • Porifera: Sponges II. Cnidaria & Ctenophora • Tissues • Symmetry I. Outline the • Germ Layers III. Lophotrochozoa unique • Embryonic characteristics Development of sponges IV. Ecdysozoa • Body Cavities • Segmentation Parazoa Parazoa • Porifera: Sponges • Porifera: Sponges – Multicellular without – Hermaphrodites tissues – Sexual and asexual reproduction – Choanocytes (collar cells) use flagella to move water and nutrients into pores – Intracellular digestion Fig. 25.11 Animal diversity, part 2 Clicker Question Diversity 2 I. Parazoa In diploblastic animals, the inner lining of the digestive cavity or tract is derived from II. Cnidaria & Ctenophora A. Endoderm. II. Outline the B. Ectoderm. unique III. Lophotrochozoa C. Mesoderm. characteristics D. Coelom. of cnidarians and IV. Ecdysozoa ctenophores 2 Coral Box jelly Cnidaria and Ctenophora • Cnidarians – Coral; sea anemone; jellyfish; hydra; box jellies • Ctenophores – Comb jellies Sea anemone Jellyfish Hydra Comb jelly Cnidaria and Ctenophora Fig. 25.12 Coral Box jelly Cnidaria and Ctenophora • Tissues Fig. 25.12 – -

Placozoans Are Eumetazoans Related to Cnidaria
bioRxiv preprint doi: https://doi.org/10.1101/200972; this version posted October 11, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Placozoans are eumetazoans related to Cnidaria Christopher E. Laumer1,2, Harald Gruber-Vodicka3, Michael G. Hadfield4, Vicki B. Pearse5, Ana Riesgo6, 4 John C. Marioni1,2,7, and Gonzalo Giribet8 1. Wellcome Trust Sanger Institute, Hinxton, CB10 1SA, United Kingdom 2. European Molecular Biology Laboratories-European Bioinformatics Institute, Hinxton, CB10 1SD, United Kingdom 8 3. Max Planck Institute for Marine Microbiology, Celsiusstraβe 1, D-28359 Bremen, Germany 4. Kewalo Marine Laboratory, Pacific Biosciences Research Center/University of Hawaiʻi at Mānoa, 41 Ahui Street, Honolulu, HI 96813, United States of America 5. University of California, Santa Cruz, Institute of Marine Sciences, 1156 High Street, Santa 12 Cruz, CA 95064, United States of America 6. The Natural History Museum, Life Sciences, Invertebrate Division Cromwell Road, London SW7 5BD, United Kingdom 7. Cancer Research UK Cambridge Institute, University of Cambridge, Li Ka Shing Centre, 16 Robinson Way, Cambridge CB2 0RE, United Kingdom 8. Museum of Comparative Zoology, Department of Organismic and Evolutionary Biology, Harvard University, 26 Oxford Street, Cambridge, MA 02138, United States of America 20 bioRxiv preprint doi: https://doi.org/10.1101/200972; this version posted October 11, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Decoding the Fossil Record of Early Lophophorates
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 1284 Decoding the fossil record of early lophophorates Systematics and phylogeny of problematic Cambrian Lophotrochozoa AODHÁN D. BUTLER ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6214 ISBN 978-91-554-9327-1 UPPSALA urn:nbn:se:uu:diva-261907 2015 Dissertation presented at Uppsala University to be publicly examined in Hambergsalen, Geocentrum, Villavägen 16, Uppsala, Friday, 23 October 2015 at 13:15 for the degree of Doctor of Philosophy. The examination will be conducted in English. Faculty examiner: Professor Maggie Cusack (School of Geographical and Earth Sciences, University of Glasgow). Abstract Butler, A. D. 2015. Decoding the fossil record of early lophophorates. Systematics and phylogeny of problematic Cambrian Lophotrochozoa. (De tidigaste fossila lofoforaterna. Problematiska kambriska lofotrochozoers systematik och fylogeni). Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 1284. 65 pp. Uppsala: Acta Universitatis Upsaliensis. ISBN 978-91-554-9327-1. The evolutionary origins of animal phyla are intimately linked with the Cambrian explosion, a period of radical ecological and evolutionary innovation that begins approximately 540 Mya and continues for some 20 million years, during which most major animal groups appear. Lophotrochozoa, a major group of protostome animals that includes molluscs, annelids and brachiopods, represent a significant component of the oldest known fossil records of biomineralised animals, as disclosed by the enigmatic ‘small shelly fossil’ faunas of the early Cambrian. Determining the affinities of these scleritome taxa is highly informative for examining Cambrian evolutionary patterns, since many are supposed stem- group Lophotrochozoa. The main focus of this thesis pertained to the stem-group of the Brachiopoda, a highly diverse and important clade of suspension feeding animals in the Palaeozoic era, which are still extant but with only with a fraction of past diversity. -

Animal Phylogeny and the Ancestry of Bilaterians: Inferences from Morphology and 18S Rdna Gene Sequences
EVOLUTION & DEVELOPMENT 3:3, 170–205 (2001) Animal phylogeny and the ancestry of bilaterians: inferences from morphology and 18S rDNA gene sequences Kevin J. Peterson and Douglas J. Eernisse* Department of Biological Sciences, Dartmouth College, Hanover NH 03755, USA; and *Department of Biological Science, California State University, Fullerton CA 92834-6850, USA *Author for correspondence (email: [email protected]) SUMMARY Insight into the origin and early evolution of the and protostomes, with ctenophores the bilaterian sister- animal phyla requires an understanding of how animal group, whereas 18S rDNA suggests that the root is within the groups are related to one another. Thus, we set out to explore Lophotrochozoa with acoel flatworms and gnathostomulids animal phylogeny by analyzing with maximum parsimony 138 as basal bilaterians, and with cnidarians the bilaterian sister- morphological characters from 40 metazoan groups, and 304 group. We suggest that this basal position of acoels and gna- 18S rDNA sequences, both separately and together. Both thostomulids is artifactal because for 1000 replicate phyloge- types of data agree that arthropods are not closely related to netic analyses with one random sequence as outgroup, the annelids: the former group with nematodes and other molting majority root with an acoel flatworm or gnathostomulid as the animals (Ecdysozoa), and the latter group with molluscs and basal ingroup lineage. When these problematic taxa are elim- other taxa with spiral cleavage. Furthermore, neither brachi- inated from the matrix, the combined analysis suggests that opods nor chaetognaths group with deuterostomes; brachiopods the root lies between the deuterostomes and protostomes, are allied with the molluscs and annelids (Lophotrochozoa), and Ctenophora is the bilaterian sister-group.