A Method for Quantifying Raga Similarity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Music Academy, Madras 115-E, Mowbray’S Road
Tyagaraja Bi-Centenary Volume THE JOURNAL OF THE MUSIC ACADEMY MADRAS A QUARTERLY DEVOTED TO THE ADVANCEMENT OF THE SCIENCE AND ART OF MUSIC Vol. XXXIX 1968 Parts MV srri erarfa i “ I dwell not in Vaikuntha, nor in the hearts of Yogins, nor in the Sun; (but) where my Bhaktas sing, there be I, Narada l ” EDITBD BY V. RAGHAVAN, M.A., p h .d . 1968 THE MUSIC ACADEMY, MADRAS 115-E, MOWBRAY’S ROAD. MADRAS-14 Annual Subscription—Inland Rs. 4. Foreign 8 sh. iI i & ADVERTISEMENT CHARGES ►j COVER PAGES: Full Page Half Page Back (outside) Rs. 25 Rs. 13 Front (inside) 20 11 Back (Do.) „ 30 „ 16 INSIDE PAGES: 1st page (after cover) „ 18 „ io Other pages (each) „ 15 „ 9 Preference will be given to advertisers of musical instruments and books and other artistic wares. Special positions and special rates on application. e iX NOTICE All correspondence should be addressed to Dr. V. Raghavan, Editor, Journal Of the Music Academy, Madras-14. « Articles on subjects of music and dance are accepted for mblication on the understanding that they are contributed solely o the Journal of the Music Academy. All manuscripts should be legibly written or preferably type written (double spaced—on one side of the paper only) and should >e signed by the writer (giving his address in full). The Editor of the Journal is not responsible for the views expressed by individual contributors. All books, advertisement moneys and cheques due to and intended for the Journal should be sent to Dr. V. Raghavan Editor. Pages. -

Syllabus for Post Graduate Programme in Music
1 Appendix to U.O.No.Acad/C1/13058/2020, dated 10.12.2020 KANNUR UNIVERSITY SYLLABUS FOR POST GRADUATE PROGRAMME IN MUSIC UNDER CHOICE BASED CREDIT SEMESTER SYSTEM FROM 2020 ADMISSION NAME OF THE DEPARTMENT: DEPARTMENT OF MUSIC NAME OF THE PROGRAMME: MA MUSIC DEPARTMENT OF MUSIC KANNUR UNIVERSITY SWAMI ANANDA THEERTHA CAMPUS EDAT PO, PAYYANUR PIN: 670327 2 SYLLABUS FOR POST GRADUATE PROGRAMME IN MUSIC UNDER CHOICE BASED CREDIT SEMESTER SYSTEM FROM 2020 ADMISSION NAME OF THE DEPARTMENT: DEPARTMENT OF MUSIC NAME OF THE PROGRAMME: M A (MUSIC) ABOUT THE DEPARTMENT. The Department of Music, Kannur University was established in 2002. Department offers MA Music programme and PhD. So far 17 batches of students have passed out from this Department. This Department is the only institution offering PG programme in Music in Malabar area of Kerala. The Department is functioning at Swami Ananda Theertha Campus, Kannur University, Edat, Payyanur. The Department has a well-equipped library with more than 1800 books and subscription to over 10 Journals on Music. We have gooddigital collection of recordings of well-known musicians. The Department also possesses variety of musical instruments such as Tambura, Veena, Violin, Mridangam, Key board, Harmonium etc. The Department is active in the research of various facets of music. So far 7 scholars have been awarded Ph D and two Ph D thesis are under evaluation. Department of Music conducts Seminars, Lecture programmes and Music concerts. Department of Music has conducted seminars and workshops in collaboration with Indira Gandhi National Centre for the Arts-New Delhi, All India Radio, Zonal Cultural Centre under the Ministry of Culture, Government of India, and Folklore Academy, Kannur. -

Navarathri Mandapam CHAPTER 4 Musical Aspect of Maharaja’S Compositions
Navarathri Mandapam CHAPTER 4 Musical Aspect of Maharaja’s Compositions 4.1. Introduction “Music begins where the possibilities of language end.” - Jean Sibelius Music is just not confined only to notes and its rendition, it is a unit of melody, its combinations and beautiful body movements. Therefore it is called Samageetam (g“rV_²) and Sharangdeva has given an apt definition to the term - JrV§ dmÚ§ VWm Z¥Ë`§, Ì`§ g“rV_wÀ`Vo& Maharaja’s compositions are models of all the three faculties of music. They are sung, played on various instruments and some compositions are exclusively composed for dance performances. To understand the nuance and technical aspects of music, it is very necessary to look back at the history of both the streams of Indian Music which are prevalent. As discussed in the earlier chapters, North Indian Music, popularly known as the Hindusthani Music had a lot of transitions since the Vedic era to the Mughal or the pre- indehendence era. After the decline of the Mughal Empire, the patronage of music continued in smaller princely kingdoms like Gwalior, Jaipur, Patiala giving rise to diversity of styles that is today known as Gharanas. Meanwhile the Bhakti and Sufi traditions -------------------------------- ( 100 ) ---------------------------------- continued to develop and interact with the different schools of music. Gharana system had a peculiar tradition of one-to-one teaching which was imparted through the Guru-Shishya tradition. To a large extent, it was limited to the palace and dance halls. It was shunned by the intellectuals, avoided by the educated middle class, and in general looked down upon as a frivolous practise. -

Rakti in Raga and Laya
VEDAVALLI SPEAKS Sangita Kalanidhi R. Vedavalli is not only one of the most accomplished of our vocalists, she is also among the foremost thinkers of Carnatic music today with a mind as insightful and uncluttered as her music. Sruti is delighted to share her thoughts on a variety of topics with its readers. Rakti in raga and laya Rakti in raga and laya’ is a swara-oriented as against gamaka- complex theme which covers a oriented raga-s. There is a section variety of aspects. Attempts have of exponents which fears that ‘been made to interpret rakti in the tradition of gamaka-oriented different ways. The origin of the singing is giving way to swara- word ‘rakti’ is hard to trace, but the oriented renditions. term is used commonly to denote a manner of singing that is of a Yo asau Dhwaniviseshastu highly appreciated quality. It swaravamavibhooshitaha carries with it a sense of intense ranjako janachittaanaam involvement or engagement. Rakti Sankarabharanam or Bhairavi? rasa raga udaahritaha is derived from the root word Tyagaraja did not compose these ‘ranj’ – ranjayati iti ragaha, ranjayati kriti-s as a cluster under the There is a reference to ‘dhwani- iti raktihi. That which is pleasing, category of ghana raga-s. Older visesha’ in this sloka from Brihaddcsi. which engages the mind joyfully texts record these five songs merely Scholars have suggested that may be called rakti. The term rakti dhwanivisesha may be taken th as Tyagaraja’ s compositions and is not found in pre-17 century not as the Pancharatna kriti-s. Not to connote sruti and that its texts like Niruktam, Vyjayanti and only are these raga-s unsuitable for integration with music ensures a Amarakosam. -

M.A-Music-Vocal-Syllabus.Pdf
BANGALORE UNIVERSITY NAAC ACCREDITED WITH ‘A’ GRADE P.G. DEPARTMENT OF PERFORMING ARTS JNANABHARATHI, BANGALORE-560056 MUSIC SYLLABUS – M.A KARNATAKA MUSIC VOCAL AND INSTRUMENTAL (VEENA, VIOLIN AND FLUTE) CBCS SYSTEM- 2014 Dr. B.M. Jayashree. Professor of Music Chairperson, BOS (PG) M.A. KARNATAKA MUSIC VOCAL AND INSTRUMENTAL (VEENA, VIOLIN AND FLUTE) Semester scheme syllabus CBCS Scheme of Examination, continuous Evaluation and other Requirements: 1. ELIGIBILITY: A Degree with music vocal/instrumental as one of the optional subject with at least 50% in the concerned optional subject an merit internal among these applicant Of A Graduate with minimum of 50% marks secured in the senior grade examination in music (vocal/instrumental) conducted by secondary education board of Karnataka OR a graduate with a minimum of 50% marks secured in PG Diploma or 2 years diploma or 4 year certificate course in vocal/instrumental music conducted either by any recognized Universities of any state out side Karnataka or central institution/Universities Any degree with: a) Any certificate course in music b) All India Radio/Doordarshan gradation c) Any diploma in music or five years of learning certificate by any veteran musician d) Entrance test (practical) is compulsory for admission. 2. M.A. MUSIC course consists of four semesters. 3. First semester will have three theory paper (core), three practical papers (core) and one practical paper (soft core). 4. Second semester will have three theory papers (core), two practical papers (core), one is project work/Dissertation practical paper and one is practical paper (soft core) 5. Third semester will have two theory papers (core), three practical papers (core) and one is open Elective Practical paper 6. -

CARNATIC MUSIC (CODE – 032) CLASS – X (Melodic Instrument) 2020 – 21 Marking Scheme
CARNATIC MUSIC (CODE – 032) CLASS – X (Melodic Instrument) 2020 – 21 Marking Scheme Time - 2 hrs. Max. Marks : 30 Part A Multiple Choice Questions: Attempts any of 15 Question all are of Equal Marks : 1. Raga Abhogi is Janya of a) Karaharapriya 2. 72 Melakarta Scheme has c) 12 Chakras 3. Identify AbhyasaGhanam form the following d) Gitam 4. Idenfity the VarjyaSwaras in Raga SuddoSaveri b) GhanDharam – NishanDham 5. Raga Harikambhoji is a d) Sampoorna Raga 6. Identify popular vidilist from the following b) M. S. Gopala Krishnan 7. Find out the string instrument which has frets d) Veena 8. Raga Mohanam is an d) Audava – Audava Raga 9. Alankaras are set to d) 7 Talas 10 Mela Number of Raga Maya MalawaGoula d) 15 11. Identify the famous flutist d) T R. Mahalingam 12. RupakaTala has AksharaKals b) 6 13. Indentify composer of Navagrehakritis c) MuthuswaniDikshitan 14. Essential angas of kriti are a) Pallavi-Anuppallavi- Charanam b) Pallavi –multifplecharanma c) Pallavi – MukkyiSwaram d) Pallavi – Charanam 15. Raga SuddaDeven is Janya of a) Sankarabharanam 16. Composer of Famous GhanePanchartnaKritis – identify a) Thyagaraja 17. Find out most important accompanying instrument for a vocal concert b) Mridangam 18. A musical form set to different ragas c) Ragamalika 19. Identify dance from of music b) Tillana 20. Raga Sri Ranjani is Janya of a) Karahara Priya 21. Find out the popular Vena artist d) S. Bala Chander Part B Answer any five questions. All questions carry equal marks 5X3 = 15 1. Gitam : Gitam are the simplest musical form. The term “Gita” means song it is melodic extension of raga in which it is composed. -
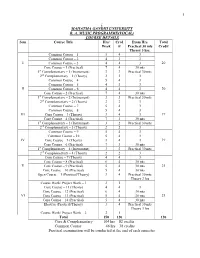
1 ; Mahatma Gandhi University B. A. Music Programme(Vocal
1 ; MAHATMA GANDHI UNIVERSITY B. A. MUSIC PROGRAMME(VOCAL) COURSE DETAILS Sem Course Title Hrs/ Cred Exam Hrs. Total Week it Practical 30 mts Credit Theory 3 hrs. Common Course – 1 5 4 3 Common Course – 2 4 3 3 I Common Course – 3 4 4 3 20 Core Course – 1 (Practical) 7 4 30 mts 1st Complementary – 1 (Instrument) 3 3 Practical 30 mts 2nd Complementary – 1 (Theory) 2 2 3 Common Course – 4 5 4 3 Common Course – 5 4 3 3 II Common Course – 6 4 4 3 20 Core Course – 2 (Practical) 7 4 30 mts 1st Complementary – 2 (Instrument) 3 3 Practical 30 mts 2nd Complementary – 2 (Theory) 2 2 3 Common Course – 7 5 4 3 Common Course – 8 5 4 3 III Core Course – 3 (Theory) 3 4 3 19 Core Course – 4 (Practical) 7 3 30 mts 1st Complementary – 3 (Instrument) 3 2 Practical 30 mts 2nd Complementary – 3 (Theory) 2 2 3 Common Course – 9 5 4 3 Common Course – 10 5 4 3 IV Core Course – 5 (Theory) 3 4 3 19 Core Course – 6 (Practical) 7 3 30 mts 1st Complementary – 4 (Instrument) 3 2 Practical 30 mts 2nd Complementary – 4 (Theory) 2 2 3 Core Course – 7 (Theory) 4 4 3 Core Course – 8 (Practical) 6 4 30 mts V Core Course – 9 (Practical) 5 4 30 mts 21 Core Course – 10 (Practical) 5 4 30 mts Open Course – 1 (Practical/Theory) 3 4 Practical 30 mts Theory 3 hrs Course Work/ Project Work – 1 2 1 Core Course – 11 (Theory) 4 4 3 Core Course – 12 (Practical) 6 4 30 mts VI Core Course – 13 (Practical) 5 4 30 mts 21 Core Course – 14 (Practical) 5 4 30 mts Elective (Practical/Theory) 3 4 Practical 30 mts Theory 3 hrs Course Work/ Project Work – 2 2 1 Total 150 120 120 Core & Complementary 104 hrs 82 credits Common Course 46 hrs 38 credits Practical examination will be conducted at the end of each semester 2 MAHATMA GANDHI UNIVERSITY B. -

Sanjay Subrahmanyan……………………………Revathi Subramony & Sanjana Narayanan
Table of Contents From the Publications & Outreach Committee ..................................... Lakshmi Radhakrishnan ............ 1 From the President’s Desk ...................................................................... Balaji Raghothaman .................. 2 Connect with SRUTI ............................................................................................................................ 4 SRUTI at 30 – Some reflections…………………………………. ........... Mani, Dinakar, Uma & Balaji .. 5 A Mellifluous Ode to Devi by Sikkil Gurucharan & Anil Srinivasan… .. Kamakshi Mallikarjun ............. 11 Concert – Sanjay Subrahmanyan……………………………Revathi Subramony & Sanjana Narayanan ..... 14 A Grand Violin Trio Concert ................................................................... Sneha Ramesh Mani ................ 16 What is in a raga’s identity – label or the notes?? ................................... P. Swaminathan ...................... 18 Saayujya by T.M.Krishna & Priyadarsini Govind ................................... Toni Shapiro-Phim .................. 20 And the Oscar goes to …… Kaapi – Bombay Jayashree Concert .......... P. Sivakumar ......................... 24 Saarangi – Harsh Narayan ...................................................................... Allyn Miner ........................... 26 Lec-Dem on Bharat Ratna MS Subbulakshmi by RK Shriramkumar .... Prabhakar Chitrapu ................ 28 Bala Bhavam – Bharatanatyam by Rumya Venkateshwaran ................. Roopa Nayak ......................... 33 Dr. M. Balamurali -

Dance Troupe
DANCE TROUPE The mission of our college ‘Blooming Humanity through Arts’ is disseminated by our institution through the program ‘Dance troupe’ globally for the past 39 years. It started on 27.12.1978 with the classical and folk dances of India and also dance dramas. The 1000th troupe programme was done on 13.01.2002 in the presence of the former Tamilnadu Chief-minister Dr.M.Kalaignar. It is also of great pride to say that soon the dance troupe will be performing its 3000thprogramme. KalaiKaviri performs dances not only to raga and thala but also performs dances with important didactic themes to reach the audience. Dances are performed with the following themes: Spirituality, Humanity, Social development, Moral values, Awareness, Religious morals and Patriotism. The troupe performs mainly in rural and remote areas to create awareness among the public. Irrespective of religion, caste and creed talented students from different backgrounds are selected, trained professionally for the troupe dances. This helps in the growth of the student’s individuality and also showcases their talent. The syllabus and dance items which students learn in their class are highly motivating for them to do a performance in the dance troupe. Main advantages of performing in a dance troupe include losing stage fear, sense of timing and rhythm, learning make-up techniques, costume selection and designing and helps building self-confidence in oneself. A student gets the opportunity to perform different roles and styles for example, classical, Indian folk and drama roles, hence by quick costume and role changes one’s concentration power increases. -

Andhra University 1 Year Diploma in (Music) Syllabus
SCHOOL OF DISTANCE EDUCATIN – ANDHRA UNIVERSITY 1st YEAR DIPLOMA IN (MUSIC) SYLLABUS (BOS approved and modified syllabus to be implemented from the admitted Batch 2012-13) Theory 100 Marks Paper I : Technical aspects of South Indian Classical Music 1. Technical Terms: a) Nada b) Sruti c) Swaras d) Swarasthanas e) Sathayi 2. Tala System: Sapta Talas, 35 Talas, Tala Dasa Prans, Chapu Tala Varieties Desadi and Madhyadi Talas 3. Musical Forms and their Lakshnas: Gitam, Varnam, Kritis, Kirtana, Padam, Ragamalika, Jasti Swaram, Swarajati, Tillana and Javali. 4. Lakshanas and Sancharas of the following Ragas: A a) Todi, b) Mayamalavagoula, c) Bhairavi, d) Kambhoji, e) Sankarabharanam f) Kalyani, g) Kharaharapriya, h) Mohana, i) Madhyamavathi j) Bilahari B. a) Dhanyasi, b) Saveri e)Vasanta d) HIndola e) Ananda Bhairavi f) Mukhari g) Kanada h) Khamas i) Begada j) Poorikalyani Paper II : Theoretical Aspects of South Indian Classical Music 100 Marks 1. Raaga and raga Lakshanam: a) Definition and Classification of ragas. b) Study of 13 Lakshanas c) ragalapana Padhathi 2. Musical Instruments and their classification 3. Special study of Tambura, Veena, Violin, Flute, Nagaswaram and Mridangam. 4. Characteristics of a composer. 5. Short biographical sketches of the following: a) Jayadeva b) Annamayya, c) Purandhara Dasa d) Narayana Tirtha e) Ramadasa f) Kshetrayya. Practical (First year) 100 Marks Paper III (Practical I) Fundamentals of Classical Music 1. a. Saraliswaras 6 b. Janta swaras 8 c. Alankaras 7 2. Gitas - 7 (Two Pillari Gitas, Two Ghanaraga Gitas, one Dhruva and one Lakshana gitam) 3. One Swarapallavi and one Swarajati 4. Five Adi Tala Varnas. -

2008 President-Elect - S
52 SRUTI BOARD OF DIRECTORS SRUTI DAY President - C. Nataraj December 2008 President-elect - S. Vidyasankar Treasurer - Venkat Kilambi Director of Resources & Development - Ramaa Nathan Director of Publications & Outreach - Vijaya Viswanathan Director of Marketing & Publicity - Srinivas Pothukuchi Director 1 - Revathi Sivakumar Director 2 Seetha Ayyalasomayajula Resource Committee - Ramaa Nathan, Uma Prabhakar, C. Nataraj, Vidyashankar Sundaresan, and Venkat Kilambi Sruti web site managed by V V Raman S R U T I The India Music & Dance Society Philadelphia, PA 2 51 Editor’s Note Welcome to our year end event, Sruti Day. This is- sue of Sruti Ranjani carries articles by some of our young- Solution to Jumble sters, who have achieved certain significant milestones in their journeys as performing artists of classical music and dance. Apart from this, our adult members have contrib- uted interesting articles that you are certain to enjoy along C O P with some puzzles and a couple of reviews of past Sruti concerts. Again, many thanks to all for taking the time to V E R S E write for this issue. H A P P Y Thanks, O D O R D E M O N S Vijaya Viswanathan 610-640-5375 C O M P O S E R’S CONTENTS DAY 1. Program - 3 2. About the Artists 3 3. Articles by members of the community 4 4. Puzzles 48 50 3 PROGRAM 2:00 PM General Body Meeting and Elections to 2009- 2010 Board (open to Sruti members only) 3:30 PM Snack Break 4:00 PM Carnatic Flute Concert by Shri V. -

Translations of Krithis of Ashok Madhava Contents
Translations of Krithis of ashok Madhava I am a multi linguist and enjoyed translating compositions of another Multi linguist. I have translated 59 of his compositions from Sanskrit, Tamil, Telugu and kannada. I enjoyed it Contents Translations of Krithis of ashok Madhava ................................................. 1 1. Abhaghi Naanalla(Kannada) ........................................................ 3 2. Adhi Nayakim(Sanskrit) ................................................................ 5 3. Amba YUvathi(Sanskrit) ............................................................. 6 4. Anbu vellame (Tamil) ................................................................... 8 5. Aravinda Nayanam (Sanskrit) ....................................................... 9 6. Arul Tharuvai shri(tamil) ........................................................... 10 7. Baaramma Hogona(Kannada) ................................................... 12 8. Bandhaa Krishna(Kannada) ....................................................... 13 9. Bhagavathi neene hari manohari (kannada).............................. 15 10. Bhajami Manasa(sanskrit) ......................................................... 16 11. Bhajamyaham satatam(sanskrit) .............................................. 18 12. Bhajana seyyu(telugu)................................................................ 20 13. BHajare re sriman(Sanskrit) ...................................................... 21 14. BHajeham sri(Sanskrit) ..............................................................