Host-Specialist Lineages Dominate the Adaptive Radiation of Reef Coral Endosymbionts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Unfolding the Secrets of Coral–Algal Symbiosis
The ISME Journal (2015) 9, 844–856 & 2015 International Society for Microbial Ecology All rights reserved 1751-7362/15 www.nature.com/ismej ORIGINAL ARTICLE Unfolding the secrets of coral–algal symbiosis Nedeljka Rosic1, Edmund Yew Siang Ling2, Chon-Kit Kenneth Chan3, Hong Ching Lee4, Paulina Kaniewska1,5,DavidEdwards3,6,7,SophieDove1,8 and Ove Hoegh-Guldberg1,8,9 1School of Biological Sciences, The University of Queensland, St Lucia, Queensland, Australia; 2University of Queensland Centre for Clinical Research, The University of Queensland, Herston, Queensland, Australia; 3School of Agriculture and Food Sciences, The University of Queensland, St Lucia, Queensland, Australia; 4The Kinghorn Cancer Centre, Garvan Institute of Medical Research, Sydney, New South Wales, Australia; 5Australian Institute of Marine Science, Townsville, Queensland, Australia; 6School of Plant Biology, University of Western Australia, Perth, Western Australia, Australia; 7Australian Centre for Plant Functional Genomics, The University of Queensland, St Lucia, Queensland, Australia; 8ARC Centre of Excellence for Coral Reef Studies, The University of Queensland, St Lucia, Queensland, Australia and 9Global Change Institute and ARC Centre of Excellence for Coral Reef Studies, The University of Queensland, St Lucia, Queensland, Australia Dinoflagellates from the genus Symbiodinium form a mutualistic symbiotic relationship with reef- building corals. Here we applied massively parallel Illumina sequencing to assess genetic similarity and diversity among four phylogenetically diverse dinoflagellate clades (A, B, C and D) that are commonly associated with corals. We obtained more than 30 000 predicted genes for each Symbiodinium clade, with a majority of the aligned transcripts corresponding to sequence data sets of symbiotic dinoflagellates and o2% of sequences having bacterial or other foreign origin. -

Checklist of Fish and Invertebrates Listed in the CITES Appendices
JOINTS NATURE \=^ CONSERVATION COMMITTEE Checklist of fish and mvertebrates Usted in the CITES appendices JNCC REPORT (SSN0963-«OStl JOINT NATURE CONSERVATION COMMITTEE Report distribution Report Number: No. 238 Contract Number/JNCC project number: F7 1-12-332 Date received: 9 June 1995 Report tide: Checklist of fish and invertebrates listed in the CITES appendices Contract tide: Revised Checklists of CITES species database Contractor: World Conservation Monitoring Centre 219 Huntingdon Road, Cambridge, CB3 ODL Comments: A further fish and invertebrate edition in the Checklist series begun by NCC in 1979, revised and brought up to date with current CITES listings Restrictions: Distribution: JNCC report collection 2 copies Nature Conservancy Council for England, HQ, Library 1 copy Scottish Natural Heritage, HQ, Library 1 copy Countryside Council for Wales, HQ, Library 1 copy A T Smail, Copyright Libraries Agent, 100 Euston Road, London, NWl 2HQ 5 copies British Library, Legal Deposit Office, Boston Spa, Wetherby, West Yorkshire, LS23 7BQ 1 copy Chadwick-Healey Ltd, Cambridge Place, Cambridge, CB2 INR 1 copy BIOSIS UK, Garforth House, 54 Michlegate, York, YOl ILF 1 copy CITES Management and Scientific Authorities of EC Member States total 30 copies CITES Authorities, UK Dependencies total 13 copies CITES Secretariat 5 copies CITES Animals Committee chairman 1 copy European Commission DG Xl/D/2 1 copy World Conservation Monitoring Centre 20 copies TRAFFIC International 5 copies Animal Quarantine Station, Heathrow 1 copy Department of the Environment (GWD) 5 copies Foreign & Commonwealth Office (ESED) 1 copy HM Customs & Excise 3 copies M Bradley Taylor (ACPO) 1 copy ^\(\\ Joint Nature Conservation Committee Report No. -

Corals of Rottnest Island Mscience Pty Ltd June 2012 Volume 1, Issue 1
Corals of Rottnest Island MScience Pty Ltd June 2012 Volume 1, Issue 1 MARINE RESEARCH Produced with the Assistance of the Rottnest Island Authority Cover picture courtesy of H. Shortland Jones * Several of the images in this publication are not from Rottnest and were sourced from corals.aims.gov.au courtesy of Dr JEN Veron Contents What are hard corals? ...........................................................................................4 Corals at Rottnest Island .....................................................................................5 Coral Identification .............................................................................................5 Hard corals found at Rottnest Island ...............................................................6 Faviidae ............................................................................................................7 Acroporidae and Pocilloporidae ....................................................................11 Dendrophyllidae ............................................................................................13 Mussidae ......................................................................................................15 Poritidae .......................................................................................................17 Siderastreidae ..............................................................................................19 Coral Identification using a customised key ................................................21 Terms used -

Biodiversity of the Kermadec Islands and Offshore Waters of the Kermadec Ridge: Report of a Coastal, Marine Mammal and Deep-Sea Survey (TAN1612)
Biodiversity of the Kermadec Islands and offshore waters of the Kermadec Ridge: report of a coastal, marine mammal and deep-sea survey (TAN1612) New Zealand Aquatic Environment and Biodiversity Report No. 179 Clark, M.R.; Trnski, T.; Constantine, R.; Aguirre, J.D.; Barker, J.; Betty, E.; Bowden, D.A.; Connell, A.; Duffy, C.; George, S.; Hannam, S.; Liggins, L..; Middleton, C.; Mills, S.; Pallentin, A.; Riekkola, L.; Sampey, A.; Sewell, M.; Spong, K.; Stewart, A.; Stewart, R.; Struthers, C.; van Oosterom, L. ISSN 1179-6480 (online) ISSN 1176-9440 (print) ISBN 978-1-77665-481-9 (online) ISBN 978-1-77665-482-6 (print) January 2017 Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries websites at: http://www.mpi.govt.nz/news-resources/publications.aspx http://fs.fish.govt.nz go to Document library/Research reports © Crown Copyright - Ministry for Primary Industries TABLE OF CONTENTS EXECUTIVE SUMMARY 1 1. INTRODUCTION 3 1.1 Objectives: 3 1.2 Objective 1: Benthic offshore biodiversity 3 1.3 Objective 2: Marine mammal research 4 1.4 Objective 3: Coastal biodiversity and connectivity 5 2. METHODS 5 2.1 Survey area 5 2.2 Survey design 6 Offshore Biodiversity 6 Marine mammal sampling 8 Coastal survey 8 Station recording 8 2.3 Sampling operations 8 Multibeam mapping 8 Photographic transect survey 9 Fish and Invertebrate sampling 9 Plankton sampling 11 Catch processing 11 Environmental sampling 12 Marine mammal sampling 12 Dive sampling operations 12 Outreach 13 3. -

Sexual Reproduction of the Solitary Sunset Cup Coral Leptopsammia Pruvoti (Scleractinia: Dendrophylliidae) in the Mediterranean
Marine Biology (2005) 147: 485–495 DOI 10.1007/s00227-005-1567-z RESEARCH ARTICLE S. Goffredo Æ J. Radetic´Æ V. Airi Æ F. Zaccanti Sexual reproduction of the solitary sunset cup coral Leptopsammia pruvoti (Scleractinia: Dendrophylliidae) in the Mediterranean. 1. Morphological aspects of gametogenesis and ontogenesis Received: 16 July 2004 / Accepted: 18 December 2004 / Published online: 3 March 2005 Ó Springer-Verlag 2005 Abstract Information on the reproduction in scleractin- came indented, assuming a sickle or dome shape. We can ian solitary corals and in those living in temperate zones hypothesize that the nucleus’ migration and change of is notably scant. Leptopsammia pruvoti is a solitary coral shape may have to do with facilitating fertilization and living in the Mediterranean Sea and along Atlantic determining the future embryonic axis. During oogene- coasts from Portugal to southern England. This coral sis, oocyte diameter increased from a minimum of 20 lm lives in shaded habitats, from the surface to 70 m in during the immature stage to a maximum of 680 lm depth, reaching population densities of >17,000 indi- when mature. Embryogenesis took place in the coelen- viduals mÀ2. In this paper, we discuss the morphological teron. We did not see any evidence that even hinted at aspects of sexual reproduction in this species. In a sep- the formation of a blastocoel; embryonic development arate paper, we report the quantitative data on the an- proceeded via stereoblastulae with superficial cleavage. nual reproductive cycle and make an interspecific Gastrulation took place by delamination. Early and late comparison of reproductive traits among Dend- embryos had diameters of 204–724 lm and 290–736 lm, rophylliidae aimed at defining different reproductive respectively. -

Volume 2. Animals
AC20 Doc. 8.5 Annex (English only/Seulement en anglais/Únicamente en inglés) REVIEW OF SIGNIFICANT TRADE ANALYSIS OF TRADE TRENDS WITH NOTES ON THE CONSERVATION STATUS OF SELECTED SPECIES Volume 2. Animals Prepared for the CITES Animals Committee, CITES Secretariat by the United Nations Environment Programme World Conservation Monitoring Centre JANUARY 2004 AC20 Doc. 8.5 – p. 3 Prepared and produced by: UNEP World Conservation Monitoring Centre, Cambridge, UK UNEP WORLD CONSERVATION MONITORING CENTRE (UNEP-WCMC) www.unep-wcmc.org The UNEP World Conservation Monitoring Centre is the biodiversity assessment and policy implementation arm of the United Nations Environment Programme, the world’s foremost intergovernmental environmental organisation. UNEP-WCMC aims to help decision-makers recognise the value of biodiversity to people everywhere, and to apply this knowledge to all that they do. The Centre’s challenge is to transform complex data into policy-relevant information, to build tools and systems for analysis and integration, and to support the needs of nations and the international community as they engage in joint programmes of action. UNEP-WCMC provides objective, scientifically rigorous products and services that include ecosystem assessments, support for implementation of environmental agreements, regional and global biodiversity information, research on threats and impacts, and development of future scenarios for the living world. Prepared for: The CITES Secretariat, Geneva A contribution to UNEP - The United Nations Environment Programme Printed by: UNEP World Conservation Monitoring Centre 219 Huntingdon Road, Cambridge CB3 0DL, UK © Copyright: UNEP World Conservation Monitoring Centre/CITES Secretariat The contents of this report do not necessarily reflect the views or policies of UNEP or contributory organisations. -

Scleractinia Fauna of Taiwan I
Scleractinia Fauna of Taiwan I. The Complex Group 台灣石珊瑚誌 I. 複雜類群 Chang-feng Dai and Sharon Horng Institute of Oceanography, National Taiwan University Published by National Taiwan University, No.1, Sec. 4, Roosevelt Rd., Taipei, Taiwan Table of Contents Scleractinia Fauna of Taiwan ................................................................................................1 General Introduction ........................................................................................................1 Historical Review .............................................................................................................1 Basics for Coral Taxonomy ..............................................................................................4 Taxonomic Framework and Phylogeny ........................................................................... 9 Family Acroporidae ............................................................................................................ 15 Montipora ...................................................................................................................... 17 Acropora ........................................................................................................................ 47 Anacropora .................................................................................................................... 95 Isopora ...........................................................................................................................96 Astreopora ......................................................................................................................99 -

The Earliest Diverging Extant Scleractinian Corals Recovered by Mitochondrial Genomes Isabela G
www.nature.com/scientificreports OPEN The earliest diverging extant scleractinian corals recovered by mitochondrial genomes Isabela G. L. Seiblitz1,2*, Kátia C. C. Capel2, Jarosław Stolarski3, Zheng Bin Randolph Quek4, Danwei Huang4,5 & Marcelo V. Kitahara1,2 Evolutionary reconstructions of scleractinian corals have a discrepant proportion of zooxanthellate reef-building species in relation to their azooxanthellate deep-sea counterparts. In particular, the earliest diverging “Basal” lineage remains poorly studied compared to “Robust” and “Complex” corals. The lack of data from corals other than reef-building species impairs a broader understanding of scleractinian evolution. Here, based on complete mitogenomes, the early onset of azooxanthellate corals is explored focusing on one of the most morphologically distinct families, Micrabaciidae. Sequenced on both Illumina and Sanger platforms, mitogenomes of four micrabaciids range from 19,048 to 19,542 bp and have gene content and order similar to the majority of scleractinians. Phylogenies containing all mitochondrial genes confrm the monophyly of Micrabaciidae as a sister group to the rest of Scleractinia. This topology not only corroborates the hypothesis of a solitary and azooxanthellate ancestor for the order, but also agrees with the unique skeletal microstructure previously found in the family. Moreover, the early-diverging position of micrabaciids followed by gardineriids reinforces the previously observed macromorphological similarities between micrabaciids and Corallimorpharia as -

CNIDARIA Corals, Medusae, Hydroids, Myxozoans
FOUR Phylum CNIDARIA corals, medusae, hydroids, myxozoans STEPHEN D. CAIRNS, LISA-ANN GERSHWIN, FRED J. BROOK, PHILIP PUGH, ELLIOT W. Dawson, OscaR OcaÑA V., WILLEM VERvooRT, GARY WILLIAMS, JEANETTE E. Watson, DENNIS M. OPREsko, PETER SCHUCHERT, P. MICHAEL HINE, DENNIS P. GORDON, HAMISH J. CAMPBELL, ANTHONY J. WRIGHT, JUAN A. SÁNCHEZ, DAPHNE G. FAUTIN his ancient phylum of mostly marine organisms is best known for its contribution to geomorphological features, forming thousands of square Tkilometres of coral reefs in warm tropical waters. Their fossil remains contribute to some limestones. Cnidarians are also significant components of the plankton, where large medusae – popularly called jellyfish – and colonial forms like Portuguese man-of-war and stringy siphonophores prey on other organisms including small fish. Some of these species are justly feared by humans for their stings, which in some cases can be fatal. Certainly, most New Zealanders will have encountered cnidarians when rambling along beaches and fossicking in rock pools where sea anemones and diminutive bushy hydroids abound. In New Zealand’s fiords and in deeper water on seamounts, black corals and branching gorgonians can form veritable trees five metres high or more. In contrast, inland inhabitants of continental landmasses who have never, or rarely, seen an ocean or visited a seashore can hardly be impressed with the Cnidaria as a phylum – freshwater cnidarians are relatively few, restricted to tiny hydras, the branching hydroid Cordylophora, and rare medusae. Worldwide, there are about 10,000 described species, with perhaps half as many again undescribed. All cnidarians have nettle cells known as nematocysts (or cnidae – from the Greek, knide, a nettle), extraordinarily complex structures that are effectively invaginated coiled tubes within a cell. -

First Records of the Sea Anemones Stichodactyla Tapetum
Turkish Journal of Zoology Turk J Zool (2015) 39: 432-437 http://journals.tubitak.gov.tr/zoology/ © TÜBİTAK Research Article doi:10.3906/zoo-1403-50 First records of the sea anemones Stichodactyla tapetum and Stichodactyla haddoni (Anthozoa: Actiniaria: Stichodactylidae) from the southeast of Iran, Chabahar (Sea of Oman) Gilan ATTARAN-FARIMAN*, Pegah JAVID Department of Marine Biology, Faculty of Marine Sciences, Chabahar Maritime University, Chabahar, Iran Received: 26.03.2014 Accepted: 28.08.2014 Published Online: 04.05.2015 Printed: 29.05.2015 Abstract: Sea anemones (order Actiniaria) are among the most widespread invertebrates in the tropical waters. The anthozoans Stichodactyla haddoni (Saville-Kent, 1893) and Stichodactyla tapetum (Hemprich & Ehrenberg in Ehrenberg, 1834) (family Stichodactylidae) were reported for the first time from the southeastern coast of Iran, Chabahar Bay, Tiss zone. The specimens of S. haddoni and S. tapetum were collected by hand from the intertidal zone of sand and rock substrates in April 2012. The samples characteristics were morphologically studied in the field and laboratory. This study presents a new locality record and information about S. haddoni and S. tapetum found in this part of the tropical sea. Key words: Exocoelic tentacles, endocoelic tentacles, tropical sea, morphological identification, symbiotic life 1. Introduction crustaceans like crabs and shrimps (Khan et al., 2004; The order Actiniaria Hertwig, 1882 (phylum Cnidaria), Katwate and Sanjeevi, 2011, Nedosyko et al., 2014), but with 46 families, includes solitary polyps with soft bodies there has been no report from Stichodactyla tapetum and nonpinnate tentacles (Daly et al., 2007). The family hosting anemonefish (Fautin et al., 2008). -

Sex, Polyps, and Medusae: Determination and Maintenance of Sex in Cnidarians†
e Reviewl Article Sex, Polyps, and Medusae: Determination and maintenance of sex in cnidarians† Runningc Head: Sex determination in Cnidaria 1* 1* i Stefan Siebert and Celina E. Juliano 1Department of Molecular and Cellular Biology, University of California, Davis, CA t 95616, USA *Correspondence may be addressed to [email protected] or [email protected] r Abbreviations:GSC, germ line stem cell; ISC, interstitial stem cell. A Keywords:hermaphrodite, gonochorism, Hydra, Hydractinia, Clytia Funding: NIH NIA 1K01AG044435-01A1, UC Davis Start Up Funds Quote:Our ability to unravel the mechanisms of sex determination in a broad array of cnidariansd requires a better understanding of the cell lineage that gives rise to germ cells. e †This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which t may lead to differences between this version and the Version of Record. Please cite this article as doi: [10.1002/mrd.22690] p e Received 8 April 2016; Revised 9 August 2016; Accepted 10 August 2016 c Molecular Reproduction & Development This article is protected by copyright. All rights reserved DOI 10.1002/mrd.22690 c This article is protected by copyright. All rights reserved A e l Abstract Mechanisms of sex determination vary greatly among animals. Here we survey c what is known in Cnidaria, the clade that forms the sister group to Bilateria and shows a broad array of sexual strategies and sexual plasticity. This observed diversity makes Cnidariai a well-suited taxon for the study of the evolution of sex determination, as closely related species can have different mechanisms, which allows for comparative studies.t In this review, we survey the extensive descriptive data on sexual systems (e.g. -
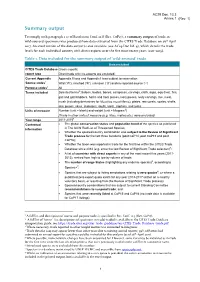
Summary Output
AC29 Doc. 13.3 Annex 1 Summary output To comply with paragraph 1 a) of Resolution Conf. 12.8 (Rev. CoP17), a summary output of trade in wild-sourced specimens was produced from data extracted from the CITES Trade Database on 26th April 2017. An excel version of the data output is also available (see AC29 Doc Inf. 4), which details the trade levels for each individual country with direct exports over the five most recent years (2011-2015). Table 1. Data included for the summary output of ‘wild-sourced’ trade Data included CITES Trade Database Gross exports; report type Direct trade only (re-exports are excluded) Current Appendix Appendix II taxa and Appendix I taxa subject to reservation Source codes1 Wild (‘W’), ranched (‘R’), unknown (‘U’) and no reported source (‘-’) Purpose codes1 All Terms included Selected terms2: baleen, bodies, bones, carapaces, carvings, cloth, eggs, egg (live), fins, gall and gall bladders, horns and horn pieces, ivory pieces, ivory carvings, live, meat, musk (including derivatives for Moschus moschiferus), plates, raw corals, scales, shells, skin pieces, skins, skeletons, skulls, teeth, trophies, and tusks. Units of measure Number (unit = blank) and weight (unit = kilogram3) [Trade in other units of measure (e.g. litres, metres etc.) were excluded] Year range 2011-20154 Contextual The global conservation status and population trend of the species as published information in The IUCN Red List of Threatened Species; Whether the species/country combination was subject to the Review of Significant Trade process for the last three iterations (post CoP14, post CoP15 and post CoP16); Whether the taxon was reported in trade for the first time within the CITES Trade Database since 2012 (e.g.