Inhibitor Development Against P7 Channel in Hepatitis C Virus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Daklinza, INN-Daclatasvir
ANNEX I SUMMARY OF PRODUCT CHARACTERISTICS 1 This medicinal product is subject to additional monitoring. This will allow quick identification of new safety information. Healthcare professionals are asked to report any suspected adverse reactions. See section 4.8 for how to report adverse reactions. 1. NAME OF THE MEDICINAL PRODUCT Daklinza 30 mg film-coated tablets Daklinza 60 mg film-coated tablets Daklinza 90 mg film-coated tablets 2. QUALITATIVE AND QUANTITATIVE COMPOSITION Daklinza 30 mg film-coated tablets Each film-coated tablet contains daclatasvir dihydrochloride equivalent to 30 mg daclatasvir. Daklinza 60 mg film-coated tablets Each film-coated tablet contains daclatasvir dihydrochloride equivalent to 60 mg daclatasvir. Daklinza 90 mg film-coated tablets Each film-coated tablet contains daclatasvir dihydrochloride equivalent to 90 mg daclatasvir. Excipient(s) with known effect Each 30-mg film-coated tablet contains 58 mg of lactose (as anhydrous). Each 60-mg film-coated tablet contains 116 mg of lactose (as anhydrous). Each 90-mg film-coated tablet contains 173 mg of lactose (as anhydrous). For the full list of excipients, see section 6.1. 3. PHARMACEUTICAL FORM Film-coated tablet (tablet). Daklinza 30 mg film-coated tablets Green biconvex pentagonal of dimensions 7.2 mm x 7.0 mm, debossed tablet with "BMS" on one side and "213" on the other side. Daklinza 60 mg film-coated tablets Light green biconvex pentagonal of dimensions 9.1 mm x 8.9 mm, debossed tablet with "BMS" on one side and "215" on the other side. Daklinza 90 mg film-coated tablets Light green biconvex round of dimension 10.16 mm diameter, embossed tablet with "BMS" on one side and "011" on the other side. -

And Ritonavir-Boosted HIV Protease Inhibitors
16 February 2012 EMA/CHMP/117973/2012 EMEA/H/C/002332/II/0004 Questions and answers on drug interactions between Victrelis (boceprevir) and ritonavir-boosted HIV protease inhibitors The European Medicines Agency has recommended changes to the prescribing information for Victrelis (boceprevir), a medicine used to treat hepatitis C, after a drug interaction study identified interactions between Victrelis and medicines used to treat HIV called ritonavir-boosted HIV protease inhibitors. These interactions could potentially reduce the effectiveness of these medicines if used together in patients being treated for both hepatitis C and HIV. The Agency’s Committee for Medicinal Products for Human Use (CHMP) has recommended the changes to ensure doctors are informed of these interactions while further data are awaited to assess the clinical impact of these drug interaction findings on these patients. What is Victrelis? Victrelis is a medicine used to treat long-term hepatitis C genotype 1 (a disease of the liver due to infection with the hepatitis C virus) in adults with compensated liver disease who have not been treated before or whose previous treatment has failed. Compensated liver disease is when the liver is damaged but is still able to work normally. Victrelis is given in combination with two other medicines, peginterferon alfa and ribavirin. The active substance in Victrelis, boceprevir, is a protease inhibitor which blocks an enzyme called HCV NS3 protease found on the hepatitis C genotype 1 virus. Victrelis was authorised in the EU in July 2011. What is the issue with Victrelis? In January 2012, the EMA was informed of the results of a study in healthy volunteers which identified drug interactions between Victrelis and the antiviral medicines atazanavir, darunavir and lopinavir, which are used to treat HIV. -

Telaprevir (Incivek)
© Hepatitis C Online PDF created September 25, 2021, 4:19 pm Telaprevir (Incivek) Discontinued. This treatment has been discontinued. Table of Contents Telaprevir Incivek Summary Drug Summary Adverse Effects Class and Mechanism Manufacturer for United States FDA Status Indications Dosing Clinical Use Cost and Medication Access Resistance Key Drug Interactions Full Prescribing Information Figures Drug Summary Although telaprevir was a promising direct-acting antiviral agent that had impact in the hepatitis C treatment field during 2011 to 2013, it was subsequently replaced by newer direct-acting antiviral agents that were more effective, better tolerated, and more convenient. Based on the dwindling role of telaprevir after newer direct-acting antiviral agents were approved, Vertex pharmaceuticals discontinued the sales and distribution of telaprevir in the United States in October 2014. Telaprevir does have some current importance since persons who previously failed a telaprevir-based regimen may have developed resistant associated variants, which could potentially impact subsequent therapy. Adverse Effects The most significant adverse effects reported in the main registration trials and in post-marketing experience were rash, anorectal complaints, and anemia. When comparing triple therapy of telaprevir, peginterferon, and ribavirin with dual therapy of peginterferon and ribavirin alone significant differences were noted with rash (56% versus 34%), anemia (36% versus 17%), and anorectal complaints that include anorectal discomfort, anal pruritus, and hemorrhoids (29% versus 7%). In most cases, the rash that develops is eczematous or maculopapular in character and mild to moderate in severity; the rash is typically manageable with good skin care and topical emollients or corticosteroids. In some instances, however, telaprevir has caused serious skin Page 1/5 rashes, including Steven's Johnson Syndrome (SJS), Drug Reaction with Eosinophilia and Systemic Symptoms (DRESS), and Toxic Epidermal Necrolysis (TEN). -

Telaprevir for HIV/Hepatitis C Virus–Coinfected Patients Failing
HIV/AIDS MAJOR ARTICLE Telaprevir for HIV/Hepatitis C Virus–Coinfected Patients Failing Treatment With Pegylated Interferon/Ribavirin (ANRS HC26 TelapreVIH): An Open-Label, Single-Arm, Phase 2 Trial Downloaded from https://academic.oup.com/cid/article/59/12/1768/2895305 by guest on 01 October 2021 Laurent Cotte,1 Joséphine Braun,2 Caroline Lascoux-Combe,3 Corine Vincent,2 Marc-Antoine Valantin,4 Philippe Sogni,5 Karine Lacombe,6 Didier Neau,7 Hugues Aumaitre,8 Dominique Batisse,9 Pierre de Truchis,10 Anne Gervais,11 Christian Michelet,12 Philippe Morlat,13 Daniel Vittecoq,14 Isabelle Rosa,15 Inga Bertucci,16 Stéphane Chevaliez,17 Jean-Pierre Aboulker,2 and Jean-Michel Molina3; for the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) HC26 Study Groupa 1Hospices Civils de Lyon, Croix-Rousse Hospital, and INSERM U1052, 2INSERM SC10-US019, Villejuif, 3Assistance Publique–Hôpitaux de Paris (AP-HP), Saint-Louis Hospital, University of Paris VII Denis Diderot, and Sorbonne Paris-Cité, INSERM U941, 4AP-HP, Pitié-Salpêtrière Hospital, and UMR-S 943, INSERM, 5AP-HP, Cochin Hospital and Paris Descartes University, INSERM U-1016, 6AP-HP, Saint-Antoine Hospital, Sorbonne Universités, UPMC University Paris 06, UMR-S1136, 7Pellegrin University Hospital, Bordeaux, 8Saint-Jean Hospital, Perpignan, 9AP-HP, Georges Pompidou European Hospital, Paris, 10AP-HP, Raymond Poincaré Hospital, Garches, 11AP-HP, Bichat-Claude Bernard Hospital, Paris, 12Pontchaillou University Hospital, Rennes, 13Saint-André University Hospital, Bordeaux, 14AP-HP, Bicètre Hospital, Le Kremlin-Bicètre, 15Créteil Hospital, 16French National Agency for Research on AIDS and Viral Hepatitis, Paris, and 17AP-HP, Henri Mondor Hospital, Créteil, France (See the Editorial Commentary by Rockstroh on pages 1777–8.) Background. -

Boceprevir (Victrelis™)
Boceprevir (Victrelis™) UTILIZATION MANAGEMENT CRITERIA DRUG CLASS: Protease Inhibitors BRAND (generic) NAMES: Victrelis (boceprevir); 200mg strength capsule FDA-APPROVED INDICATIONS Victrelis (boceprevir) is a hepatitis C virus (HCV) NS3/4A protease inhibitor indicated for the treatment of chronic hepatitis C (CHC) genotype 1 infection, in combination with peginterferon alfa and ribavirin, in adult patients (18 years of age or older) with compensated liver disease, including cirrhosis, who are previously untreated or who have failed previous interferon and ribavirin therapy, including prior null responders, partial responders, and relapsers. Victrelis must not be used as monotherapy and should only be used in combination with peginterferon alfa and ribavirin. The efficacy of Victrelis has not been studies in patients who have previously failed therapy with a treatment regimen that includes Victrelis or other HCV NS3/4A protease inhibitors. COVERAGE AUTHORIZATION CRITERIA INITIAL THERAPY Boceprevir (Victrelis) may be eligible for coverage when the following criteria are met: 1. The patient is 18 years of age or older; AND 2. The patient has a diagnosis of chronic hepatitis C (CHC) infection with confirmed genotype 1; AND 3. The patient: Has F2 or higher on the IASL, Batts-Ludwig, or Metavir fibrosis staging scales (medical record documentation required); OR Has F3 or higher on the Ishak fibrosis staging scale (medical record documentation required); OR Has cirrhosis secondary to CHC [Metavir F4, Ishak F5-6, or radiographic evidence of portal hypertension, esophageal varices, ascites (medical record documentation required)]; AND 4. The patient has contraindications to a sofosbuvir-based regimen (i.e., Sovaldi or Harvoni), in addition to Viekira Pak. -

Effectiveness and Safety of Daclatasvir/Sofosbuvir with Or Without Ribavirin in Genotype 3 Hepatitis C Virus Infected Patients
©The Author 2019. Published by Sociedad Española de Quimioterapia. This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0)(https://creativecommons.org/licenses/by-nc/4.0/). Original Luis Margusino-Framiñán1,5 Purificación Cid-Silva1,5 Effectiveness and safety of daclatasvir/sofosbuvir with Álvaro Mena-de-Cea2,5 Iria Rodríguez-Osorio2,5 or without ribavirin in genotype 3 hepatitis C virus Berta Pernas-Souto2,5 infected patients. Results in real clinical practice Manuel Delgado-Blanco3,5 Sonia Pertega-Díaz4 1 Isabel Martín-Herranz 1 2,5 Pharmacy Service. Universitary Hospital of A Coruña (CHUAC). Spain. Ángeles Castro-Iglesias 2Infectious Diseases Unit. Internal Medicine Service. Universitary Hospital of A Coruña (CHUAC). Spain. 3Hepatology Unit. Digestive System Service. Universitary Hospital of A Coruña (CHUAC). Spain. 4Epidemiologial Unit. Universitary Hospital of A Coruña (CHUAC). Spain. 5Division of Clinical Virology, Biomedical Research Institute of A Coruña (INIBIC), Universitary Hospital of A Coruña (CHUAC), SERGAS, University of A Coruña (UDC), A Coruña, Spain. Article history Received: 15 October 2018; Revision Requested: 8 November 2018; Revision Received: 26 November 2018; Accepted: 9 January 2019 ABSCTRACT Efectividad y seguridad de daclatasvir/ sofosbuvir con o sin ribavirina en pacientes Objectives. Direct-acting antivirals have shown high ef- infectados por el genotipo 3 del virus de la ficacy in all hepatitis C virus (HCV) genotypes, but genotype 3 hepatitis C. Resultados en práctica clínica real (G3) treatments continue to be a challenge, mainly in cirrhotic patients. The aim of this study is to analyse effectiveness and Objetivos. Los antivirales de acción directa han demostra- safety of daclatasvir associated with sofosbuvir with or without do una alta eficacia en todos los genotipos del virus de la he- ribavirin in G3-HCV infected patients in real clinical practice. -

The Legally Binding Text Is the Original French Version TRANSPARENCY
The legally binding text is the original French version TRANSPARENCY COMMITTEE OPINION 14 December 2011 INCIVO 375 mg, film-coated tablet B/4 bottles of 42 tablets (CIP code: 217 378-5) B/1 bottle of 42 tablets (CIP code: 219 249-8) Applicant: JANSSEN-CILAG telaprevir ATC Code: J05AE11 (protease inhibitor) List I Hospital prescription restricted to gastroenterology, hepatology, internal medicine or infectiology specialists. Date of Marketing Authorisation (centralised European procedure): Box of 4 bottles: 19/09/2011 Box of 1 bottle: 13/10/2011 Reason for request: Inclusion on the list of medicines refundable by National Health Insurance and on the list of medicines approved for hospital use. Medical, Economic and Public Health Assessment Division 1/22 1. CHARACTERISTICS OF THE MEDICINAL PRODUCT 1.1. Active ingredient telaprevir 1.2. Background This is an NS3/4A protease inhibitor of the genotype 1 hepatitis C virus. 1.3. Indication “INCIVO, in combination with peginterferon alfa and ribavirin, is indicated for the treatment of genotype 1 chronic hepatitis C in adult patients with compensated liver disease (including cirrhosis): - who are treatment-naive; - who have previously been treated with interferon alfa (pegylated or non-pegylated) alone or in combination with ribavirin, including relapsers, partial responders and null responders (see sections 4.4 and 5.1 of the SPC).” 1.4. Dosage “Treatment with INCIVO must be initiated and monitored by a physician experienced in the management of chronic hepatitis C. INCIVO, 750 mg dose of INCIVO (two 375 mg film-coated tablets) should be taken orally every 8 hours with food (the total daily dose is 6 tablets (2,250 mg)). -

Caracterización Molecular Del Perfil De Resistencias Del Virus De La
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Programa de doctorado en Medicina Departamento de Medicina Facultad de Medicina Universidad Autónoma de Barcelona TESIS DOCTORAL Caracterización molecular del perfil de resistencias del virus de la hepatitis C después del fallo terapéutico a antivirales de acción directa mediante secuenciación masiva Tesis para optar al grado de doctor de Qian Chen Directores de la Tesis Dr. Josep Quer Sivila Dra. Celia Perales Viejo Dr. Josep Gregori i Font Laboratorio de Enfermedades Hepáticas - Hepatitis Víricas Vall d’Hebron Institut de Recerca (VHIR) Barcelona, 2018 ABREVIACIONES Abreviaciones ADN: Ácido desoxirribonucleico AK: Adenosina quinasa ALT: Alanina aminotransferasa ARN: Ácido ribonucleico ASV: Asunaprevir BOC: Boceprevir CCD: Charge Coupled Device CLDN1: Claudina-1 CHC: Carcinoma hepatocelular DAA: Antiviral de acción directa DC-SIGN: Dendritic cell-specific ICAM-3 grabbing non-integrin DCV: Daclatasvir DSV: Dasabuvir -
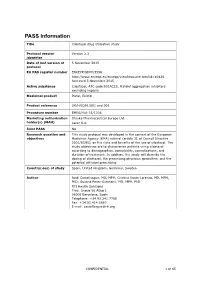
PASS Information
PASS Information Title Cilostazol drug utilisation study Protocol version Version 2.3 identifier Date of last version of 5 November 2015 protocol EU PAS register number ENCEPP/SDPP/3596 http://www.encepp.eu/encepp/viewResource.htm?id=10426 Accessed 5 November 2015 Active substance Cilostazol, ATC code B01AC23, Platelet aggregation inhibitors excluding heparin Medicinal product Pletal, Ekistol Product reference UK/H/0291/001 and 002 Procedure number EMEA/H/A-31/1306 Marketing authorisation Otsuka Pharmaceutical Europe Ltd. holder(s) (MAH) Lacer S.A. Joint PASS No Research question and This study protocol was developed in the context of the European objectives Medicines Agency (EMA) referral (article 31 of Council Directive 2001/83/EC) on the risks and benefits of the use of cilostazol. The study objectives are to characterise patients using cilostazol according to demographics, comorbidity, comedications, and duration of treatment. In addition, the study will describe the dosing of cilostazol, the prescribing physician specialties, and the potential off-label prescribing. Country(-ies) of study Spain, United Kingdom, Germany, Sweden Author Jordi Castellsague, MD, MPH; Cristina Varas-Lorenzo, MD, MPH, PhD; Susana Perez-Gutthann, MD, MPH, PhD RTI Health Solutions Trav. Gracia 56 Atico 1 08006 Barcelona, Spain Telephone: +34.93.241.7766 Fax: +34.93.414.2610 E-mail: [email protected] CONFIDENTIAL 1 of 65 Marketing Authorisation Holder(s) Marketing authorisation Otsuka Pharmaceutical Europe Ltd. holder(s) Gallions Wexham Springs Framewood Road Wexham SL3 6PJ, UK Lacer S.A. Sardenya 350 08025 Barcelona Spain MAH contact person Dr Marco Avila Regional Vice President, Medical Europe Otsuka Pharmaceutical Europe Ltd EU QPPV Dr Achint Kumar Otsuka Europe Development and Commercialisation Limited (OEDC) Head of Medical Dr Sanjay Kapoor Compliance-Europe Otsuka Pharmaceutical Europe Ltd. -

Estonian Statistics on Medicines 2016 1/41
Estonian Statistics on Medicines 2016 ATC code ATC group / Active substance (rout of admin.) Quantity sold Unit DDD Unit DDD/1000/ day A ALIMENTARY TRACT AND METABOLISM 167,8985 A01 STOMATOLOGICAL PREPARATIONS 0,0738 A01A STOMATOLOGICAL PREPARATIONS 0,0738 A01AB Antiinfectives and antiseptics for local oral treatment 0,0738 A01AB09 Miconazole (O) 7088 g 0,2 g 0,0738 A01AB12 Hexetidine (O) 1951200 ml A01AB81 Neomycin+ Benzocaine (dental) 30200 pieces A01AB82 Demeclocycline+ Triamcinolone (dental) 680 g A01AC Corticosteroids for local oral treatment A01AC81 Dexamethasone+ Thymol (dental) 3094 ml A01AD Other agents for local oral treatment A01AD80 Lidocaine+ Cetylpyridinium chloride (gingival) 227150 g A01AD81 Lidocaine+ Cetrimide (O) 30900 g A01AD82 Choline salicylate (O) 864720 pieces A01AD83 Lidocaine+ Chamomille extract (O) 370080 g A01AD90 Lidocaine+ Paraformaldehyde (dental) 405 g A02 DRUGS FOR ACID RELATED DISORDERS 47,1312 A02A ANTACIDS 1,0133 Combinations and complexes of aluminium, calcium and A02AD 1,0133 magnesium compounds A02AD81 Aluminium hydroxide+ Magnesium hydroxide (O) 811120 pieces 10 pieces 0,1689 A02AD81 Aluminium hydroxide+ Magnesium hydroxide (O) 3101974 ml 50 ml 0,1292 A02AD83 Calcium carbonate+ Magnesium carbonate (O) 3434232 pieces 10 pieces 0,7152 DRUGS FOR PEPTIC ULCER AND GASTRO- A02B 46,1179 OESOPHAGEAL REFLUX DISEASE (GORD) A02BA H2-receptor antagonists 2,3855 A02BA02 Ranitidine (O) 340327,5 g 0,3 g 2,3624 A02BA02 Ranitidine (P) 3318,25 g 0,3 g 0,0230 A02BC Proton pump inhibitors 43,7324 A02BC01 Omeprazole -

Pegasys®) Peginterferon Alfa-2B (Peg-Intron®
Peginterferon alfa-2a (Pegasys®) Peginterferon alfa-2b (Peg-Intron®) UTILIZATION MANAGEMENT CRITERIA DRUG CLASS: Pegylated Interferons BRAND (generic) NAME: Pegasys (peginterferon alfa-2a) Single-use vial 180 mcg/1.0 mL; Prefilled syringe 180 mcg/0.5 mL Peg-Intron (peginterferon alfa-2b) Single-use vial (with 1.25 mL diluent) and REDIPEN®: 50 mcg, 80 mcg, 120 mcg, 150 mcg per 0.5 mL. FDA-APPROVED INDICATIONS PEG-Intron (peginterferon alfa-2b): Combination therapy with ribavirin: • Chronic Hepatitis C (CHC) in patients 3 years of age and older with compensated liver disease. • Patients with the following characteristics are less likely to benefit from re-treatment after failing a course of therapy: previous nonresponse, previous pegylated interferon treatment, significant bridging fibrosis or cirrhosis, and genotype 1 infection. Monotherapy: CHC in patients (18 years of age and older) with compensated liver disease previously untreated with interferon alpha. Pegasys (Peginterferon alfa-2a): • Treatment of Chronic Hepatitis C (CHC) in adults with compensated liver disease not previously treated with interferon alpha, in patients with histological evidence of cirrhosis and compensated liver disease, and in adults with CHC/HIV coinfection and CD4 count > 100 cells/mm3. o Combination therapy with ribavirin is recommended unless patient has contraindication to or significant intolerance to ribavirin. Pegasys monotherapy is indicated for: • Treatment of adult patients with HBeAg positive and HBeAg negative chronic hepatitis B who have compensated -

Peginterferon, and Ribavirin Patients Treated with Daclatasvir, Hepatitis
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Hiroshima University Institutional Repository Ultradeep Sequencing Study of Chronic Hepatitis C Virus Genotype 1 Infection in Patients Treated with Daclatasvir, Peginterferon, and Ribavirin Eisuke Murakami, Michio Imamura, C. Nelson Hayes, Hiromi Abe, Nobuhiko Hiraga, Yoji Honda, Atsushi Ono, Keiichi Kosaka, Tomokazu Kawaoka, Masataka Tsuge, Hiroshi Aikata, Shoichi Takahashi, Daiki Miki, Hidenori Ochi, Hirotaka Matsui, Akinori Kanai, Toshiya Inaba, Fiona McPhee and Kazuaki Chayama Antimicrob. Agents Chemother. 2014, 58(4):2105. DOI: 10.1128/AAC.02068-13. Downloaded from Published Ahead of Print 27 January 2014. Updated information and services can be found at: http://aac.asm.org/content/58/4/2105 http://aac.asm.org/ These include: REFERENCES This article cites 35 articles, 6 of which can be accessed free at: http://aac.asm.org/content/58/4/2105#ref-list-1 CONTENT ALERTS Receive: RSS Feeds, eTOCs, free email alerts (when new articles cite this article), more» on June 25, 2014 by guest Information about commercial reprint orders: http://journals.asm.org/site/misc/reprints.xhtml To subscribe to to another ASM Journal go to: http://journals.asm.org/site/subscriptions/ Ultradeep Sequencing Study of Chronic Hepatitis C Virus Genotype 1 Infection in Patients Treated with Daclatasvir, Peginterferon, and Ribavirin Eisuke Murakami,a,b Michio Imamura,a,b C. Nelson Hayes,a,b Hiromi Abe,a,b Nobuhiko Hiraga,a,b Yoji Honda,a,b Atsushi Ono,a,b Keiichi Kosaka,a,b