Potential and Pitfalls of Low Coverage Genome Surveys for Evolutionary Biology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Classification of Living and Fossil Genera of Decapod Crustaceans
RAFFLES BULLETIN OF ZOOLOGY 2009 Supplement No. 21: 1–109 Date of Publication: 15 Sep.2009 © National University of Singapore A CLASSIFICATION OF LIVING AND FOSSIL GENERA OF DECAPOD CRUSTACEANS Sammy De Grave1, N. Dean Pentcheff 2, Shane T. Ahyong3, Tin-Yam Chan4, Keith A. Crandall5, Peter C. Dworschak6, Darryl L. Felder7, Rodney M. Feldmann8, Charles H. J. M. Fransen9, Laura Y. D. Goulding1, Rafael Lemaitre10, Martyn E. Y. Low11, Joel W. Martin2, Peter K. L. Ng11, Carrie E. Schweitzer12, S. H. Tan11, Dale Tshudy13, Regina Wetzer2 1Oxford University Museum of Natural History, Parks Road, Oxford, OX1 3PW, United Kingdom [email protected] [email protected] 2Natural History Museum of Los Angeles County, 900 Exposition Blvd., Los Angeles, CA 90007 United States of America [email protected] [email protected] [email protected] 3Marine Biodiversity and Biosecurity, NIWA, Private Bag 14901, Kilbirnie Wellington, New Zealand [email protected] 4Institute of Marine Biology, National Taiwan Ocean University, Keelung 20224, Taiwan, Republic of China [email protected] 5Department of Biology and Monte L. Bean Life Science Museum, Brigham Young University, Provo, UT 84602 United States of America [email protected] 6Dritte Zoologische Abteilung, Naturhistorisches Museum, Wien, Austria [email protected] 7Department of Biology, University of Louisiana, Lafayette, LA 70504 United States of America [email protected] 8Department of Geology, Kent State University, Kent, OH 44242 United States of America [email protected] 9Nationaal Natuurhistorisch Museum, P. O. Box 9517, 2300 RA Leiden, The Netherlands [email protected] 10Invertebrate Zoology, Smithsonian Institution, National Museum of Natural History, 10th and Constitution Avenue, Washington, DC 20560 United States of America [email protected] 11Department of Biological Sciences, National University of Singapore, Science Drive 4, Singapore 117543 [email protected] [email protected] [email protected] 12Department of Geology, Kent State University Stark Campus, 6000 Frank Ave. -

49 a New Genus and Species of Tree-Climbing Crab
THE RAFFLES BULLETIN OF ZOOLOGY 2003 THE RAFFLES BULLETIN OF ZOOLOGY 2003 51(1): 49-59 © National University of Singapore A NEW GENUS AND SPECIES OF TREE-CLIMBING CRAB (CRUSTACEA: BRACHYURA: SESARMIDAE) FROM TAIWAN WITH NOTES ON ITS ECOLOGY AND LARVAL MORPHOLOGY Christoph D. Schubart Biologie 1 (Zoologie), Universität Regensburg, 93040 Regensburg, Germany Email: [email protected] Hung-Chang Liu Institute of Zoology, Academia Sinica, Nankang, Taipei 115 Taiwan, R.O.C. José A. Cuesta Instituto de Ciencias Marinas de Andalucía (CSIC), Campus Río San Pedro, Apdo. Oficial, 11510 Puerto Real, Cádiz, Spain ABSTRACT. – A new genus and species of sesarmid crab, Scandarma lintou, is described from Taiwan. This crab has a semi-terrestrial habit: adults and juveniles thrive in wind-protected and vegetated habitats in close vicinity of fresh water and up to one kilometer from the sea. In southern Taiwan, this species was most commonly found hiding in leaf axils or climbing on the thorny leaves of the screw pine Pandanus odoratissimus Linnaeus. During the reproductive season (June to January), ovigerous females migrate to estuaries, where small and free-swimming pelagic larvae are released into the brackish waters and probably washed into the sea. Morphologically, this species is superficially similar to species of other semi-terrestrial sesarmid genera, but differs from these taxa by the shape of the anterolateral carapace region, by the markedly flattened fingers, by the presence of a row of tubercles on the dorsal border of the dactylus, a row of ventral spines on the pollex, and a granular ridge on the dorsal face of the palm. -

Molecular Phylogeny, Taxonomy, and Evolution of Nonmarine Lineages Within the American Grapsoid Crabs (Crustacea: Brachyura) Christoph D
Molecular Phylogenetics and Evolution Vol. 15, No. 2, May, pp. 179–190, 2000 doi:10.1006/mpev.1999.0754, available online at http://www.idealibrary.com on Molecular Phylogeny, Taxonomy, and Evolution of Nonmarine Lineages within the American Grapsoid Crabs (Crustacea: Brachyura) Christoph D. Schubart*,§, Jose´ A. Cuesta†, Rudolf Diesel‡, and Darryl L. Felder§ *Fakulta¨tfu¨ r Biologie I: VHF, Universita¨ t Bielefeld, Postfach 100131, 33501 Bielefeld, Germany; †Departamento de Ecologı´a,Facultad de Biologı´a,Universidad de Sevilla, Apdo. 1095, 41080 Sevilla, Spain; ‡Max-Planck-Institut fu¨ r Verhaltensphysiologie, Postfach 1564, 82305 Starnberg, Germany; and §Department of Biology and Laboratory for Crustacean Research, University of Louisiana at Lafayette, Lafayette, Louisiana 70504-2451 Received January 4, 1999; revised November 9, 1999 have attained lifelong independence from the sea (Hart- Grapsoid crabs are best known from the marine noll, 1964; Diesel, 1989; Ng and Tan, 1995; Table 1). intertidal and supratidal. However, some species also The Grapsidae and Gecarcinidae have an almost inhabit shallow subtidal and freshwater habitats. In worldwide distribution, being most predominant and the tropics and subtropics, their distribution even species rich in subtropical and tropical regions. Over- includes mountain streams and tree tops. At present, all, there are 57 grapsid genera with approximately 400 the Grapsoidea consists of the families Grapsidae, recognized species (Schubart and Cuesta, unpubl. data) Gecarcinidae, and Mictyridae, the first being subdi- and 6 gecarcinid genera with 18 species (Tu¨ rkay, 1983; vided into four subfamilies (Grapsinae, Plagusiinae, Tavares, 1991). The Mictyridae consists of a single Sesarminae, and Varuninae). To help resolve phyloge- genus and currently 4 recognized species restricted to netic relationships among these highly adaptive crabs, portions of the mitochondrial genome corresponding the Indo-West Pacific (P. -
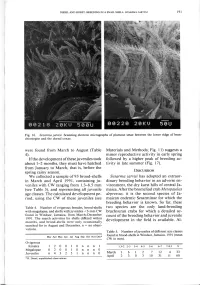
(Table 4). If the Development of These Juveniles Took About 1-2 Months
DIESEL AND HORST: BREEDING IN A SNAIL SHELL: SESARMA JARVISI 191 Fig. 16. Sesarma jarvisi. Scanning electron micrographs of plumose setae between the lower ridge of bran- chiostegite and the dorsal coxae. were found from March to August (Table Materials and Methods; Fig. 11) suggests a 4). minor reproductive activity in early spring If the development of these juveniles took followed by a higher peak of breeding ac- about 1-2 months, they must have hatched tivity in late summer (Fig. 17). from January to March, that is, before the spring rainy season. DISCUSSION We collected a sample of 93 brood-shells Sesarma jarvisi has adopted an extraor- in March and April 1991, containing ju- dinary breeding behavior in an adverse en- veniles with CW ranging from 1.3-8.5 mm vironment, the dry karst hills of central Ja- (see Table 5), and representing all juvenile maica. After the bromeliad crab Metopaulias age classes. The calculated development pe- depressus, it is the second species of Ja- riod, using the CW of these juveniles (see maican endemic Sesarminae for which the breeding behavior is known. So far, these Table 4. Number of ovigerous females, brood-shells two species are the only land-breeding with megalopae, and shells with juveniles <3-mm CW brachyuran crabs for which a detailed ac- found in Windsor, Jamaica, from March-December count of the breeding behavior and juvenile 1991. The search activities for shells differed within months, and brood-shells were only occasionally development in the field is available. Al- searched for in August and December, n = no obser- vations. -

Duffy 2010 Encyclopedia of Animal Behavior-1.Pdf
This article was originally published in the Encyclopedia of Animal Behavior published by Elsevier, and the attached copy is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non- commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues who you know, and providing a copy to your institution’s administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution’s website or repository, are prohibited. For exceptions, permission may be sought for such use through Elsevier's permissions site at: http://www.elsevier.com/locate/permissionusematerial Duffy J.E. (2010) Crustacean Social Evolution. In: Breed M.D. and Moore J., (eds.) Encyclopedia of Animal Behavior, volume 1, pp. 421-429 Oxford: Academic Press. © 2010 Elsevier Ltd. All rights reserved. Author's personal copy Crustacean Social Evolution J. E. Duffy, Virginia Institute of Marine Science, Gloucester Point, VA, USA ã 2010 Elsevier Ltd. All rights reserved. Introduction shed periodically during growth. Each of the segments in the primitive ancestral crustacean body bore a pair of The Crustacea represent one of the most spectacular appendages, which have been modified during the evolu- evolutionary radiations in the animal kingdom, whether tion of the various crustacean groups into a wide range of measured by species richness or diversity in morphology structures used in feeding, locomotion, sensation, and or lifestyles. Its members range from microscopic mites of communication. -

Generating Genomic Resources for Two Crustacean Species and Their Application to the Study of White Spot Disease
Generating genomic resources for two crustacean species and their application to the study of White Spot Disease Submitted by Bas Verbruggen To the University of Exeter as a thesis for the degree of Doctor of Philosophy in Biological Sciences, September 2016 This thesis is available for Library use on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement I certify that all material in this thesis which is not my own work has been identified and that no material has previously been submitted and approved for the award of a degree by this or any other University. Signature: ……………………………………(Bas Verbruggen) Abstract Over the last decades the crustacean aquaculture sector has been steadily growing, in order to meet global demands for its products. A major hurdle for further growth of the industry is the prevalence of viral disease epidemics that are facilitated by the intense culture conditions. A devastating virus impacting on the sector is the White Spot Syndrome Virus (WSSV), responsible for over US $ 10 billion in losses in shrimp production and trade. The Pathogenicity of WSSV is high, reaching 100 % mortality within 3-10 days in penaeid shrimps. In contrast, the European shore crab Carcinus maenas has been shown to be relatively resistant to WSSV. Uncovering the basis of this resistance could help inform on the development of strategies to mitigate the WSSV threat. C. maenas has been used widely in studies on ecotoxicology and host-pathogen interactions. However, like most aquatic crustaceans, the genomic resources available for this species are limited, impairing experimentation. -

Download Preprint
Biological Reviews Macroecology of parent al care in arthropods: higher mortality risk leads to higher benefits of offspring protection in tropical climates Journal:For Biological Review Reviews Only Manuscript ID BRV-04-2016-0076.R2 Manuscript Type: Original Article Date Submitted by the Author: n/a Complete List of Authors: Santos, Eduardo; Universidade de Sao Paulo, Departamento de Zoologia Bueno, Pedro; Universidade de Sao Paulo, Departamento de Ecologia Gilbert, James; University of Hull, School of Biological, Biomedical & Environmental Sciences Machado, Glauco; Universidade de Sao Paulo, Departamento de Ecologia abiotic factors, biotic interactions, evapotranspiration, egg attendance, egg Keywords: coating, meta-regression, nest, parasitism, parental removal, predation Page 1 of 60 Biological Reviews 1 2 3 1 Macroecology of parental care in arthropods: higher mortality 4 5 6 2 risk leads to higher benefits of offspring protection in tropical 7 8 9 3 climates 10 11 12 4 13 14 5 Eduardo S. A. Santos 1, 2, *, Pedro P. Bueno 1, James D. J. Gilbert 3 and Glauco 15 16 1 17 6 Machado 18 For Review Only 19 7 20 21 8 1LAGE do Departamento de Ecologia, Instituto de Biociências, Universidade de São Paulo, 22 23 24 9 Rua do Matão, trav. 14, no 321, Cidade Universitária, 05508-090, São Paulo, SP, Brazil 25 2 26 10 BECO do Departamento de Zoologia, Instituto de Biociências, Universidade de São Paulo, 27 28 11 Rua do Matão, trav. 14, no 321, Cidade Universitária, 05508-090, São Paulo, SP, Brazil 29 30 12 3School of Environmental Sciences, University of Hull, Cottingham Rd, Hull HU6 7RX, UK 31 32 33 13 34 35 14 36 37 15 Running title : Macroecology of parental care in arthropods 38 39 16 40 41 17 42 43 * 44 18 Author for correspondence (E-mail: [email protected]; Tel.: +55 (11) 3091-0989). -

Decapod Crustacean Phylogenetics
CRUSTACEAN ISSUES ] 3 II %. m Decapod Crustacean Phylogenetics edited by Joel W. Martin, Keith A. Crandall, and Darryl L. Felder £\ CRC Press J Taylor & Francis Group Decapod Crustacean Phylogenetics Edited by Joel W. Martin Natural History Museum of L. A. County Los Angeles, California, U.S.A. KeithA.Crandall Brigham Young University Provo,Utah,U.S.A. Darryl L. Felder University of Louisiana Lafayette, Louisiana, U. S. A. CRC Press is an imprint of the Taylor & Francis Croup, an informa business CRC Press Taylor & Francis Group 6000 Broken Sound Parkway NW, Suite 300 Boca Raton, Fl. 33487 2742 <r) 2009 by Taylor & Francis Group, I.I.G CRC Press is an imprint of 'Taylor & Francis Group, an In forma business No claim to original U.S. Government works Printed in the United States of America on acid-free paper 109 8765 43 21 International Standard Book Number-13: 978-1-4200-9258-5 (Hardcover) Ibis book contains information obtained from authentic and highly regarded sources. Reasonable efforts have been made to publish reliable data and information, but the author and publisher cannot assume responsibility for the valid ity of all materials or the consequences of their use. The authors and publishers have attempted to trace the copyright holders of all material reproduced in this publication and apologize to copyright holders if permission to publish in this form has not been obtained. If any copyright material has not been acknowledged please write and let us know so we may rectify in any future reprint. Except as permitted under U.S. Copyright Faw, no part of this book maybe reprinted, reproduced, transmitted, or uti lized in any form by any electronic, mechanical, or other means, now known or hereafter invented, including photocopy ing, microfilming, and recording, or in any information storage or retrieval system, without written permission from the publishers. -

Genetic Diversity of Freshwater Crabs (Brachyura: Sesarmidae) from Central Jamaica with Description of a New Species
Journal of Natural History, 2005; 39(6): 469–481 Genetic diversity of freshwater crabs (Brachyura: Sesarmidae) from central Jamaica with description of a new species CHRISTOPH D. SCHUBART & PETER KOLLER Fakulta¨t fu¨r Biologie 1 (Zoologie), Universita¨t Regensburg, 93040 Regensburg, Germany. E-mail: [email protected] (Accepted 2 February 2004) Abstract Jamaica is the only island of the Greater Antilles where freshwater streams are not populated by representatives of the old lineage of freshwater crabs, the Pseudothelphusidae. Instead, a very diverse fauna of endemic sesarmid crabs inhabits freshwater and terrestrial habitats throughout the island, thereby showing complete independence from the sea. They have been reported thriving in bromeliad leaf axils, rock rubble, empty snail shells, caves and mountain streams. Otherwise, the Sesarmidae are typical inhabitants of soft-sediment littoral habitats like marshes and mangroves. For many years, crabs from Jamaican mountains streams had been considered to belong to a single species, Sesarma bidentatum Benedict 1892. However, recent morphological and molecular studies have shown that crabs from mountain streams of different regions of the island belong to different species. Consequently, four new crab species have been described over the last 10 years. In this study, we give evidence that freshwater streams in central Jamaica also host two different species of crabs. In streams draining to the north we exclusively found the species S. windsor Tu¨rkay and Diesel, 1994, while streams draining to the south were inhabited by a closely related but undescribed species of crab. The southern species is here described as new and is referred to as Sesarma meridies sp. -

Parental Care in the Freshwater Crab Sylviocarcinus Pictus (Milne-Edwards, 1853)*
Vol.3, No.2, 161-163 (2013) Open Journal of Ecology http://dx.doi.org/10.4236/oje.2013.32019 Parental care in the freshwater crab Sylviocarcinus * pictus (Milne-Edwards, 1853) Bruno Sampaio Sant’Anna, Erico Luis Hoshiba Takahashi, Gustavo Yomar Hattori Federal University of Amazonas (Amazonas University), Institute of Exact Science and Technology (ICET), Itacoatiara, Brazil; *Corresponding Author: [email protected], [email protected], [email protected] Received 16 January 2013; revised 17 February 2013; accepted 15 March 2013 Copyright © 2013 Bruno Sampaio Sant’Anna et al. This is an open access article distributed under the Creative Commons Attribu- tion License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT of the bromeliad Aechmea nudicaulis (Linnaeus) Grise- bach, 1864 [6]. Jawed Hirudinidae deposit desiccation- Parental care is a common strategy in many resistant cocoons on land and many species brood the animal groups, to increase survival of the off- eggs and young [7]. spring. Here, we report parental care in the In decapod crustaceans, parental care is usually re- freshwater crab Sylviocarcinus pictus. A female stricted to females that carry the eggs in the brood com- caught in the Amazon River, Brazil, bore juvenile partment, and care is terminated when the larvae are re- crabs rather than eggs on her abdomen. Kept in leased into the plankton [8,9]. However, among other the laboratory, the female retained the juveniles examples in crustaceans, [10] presented evidence of a on the abdomen for 17 days, after which the direct link between active brood care and provision of juveniles left the abdomen. -

From Symmetry to Asymmetry
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 14279–14286, December 1996 Colloquium Paper This paper was presented at a colloquium entitled ‘‘Symmetries Throughout the Sciences,’’ organized by Ernest M. Henley, held May 11–12, 1996, at the National Academy of Sciences in Irvine, CA. From symmetry to asymmetry: Phylogenetic patterns of asymmetry variation in animals and their evolutionary significance (morphologyydevelopmentyhandednessyvertebrateyinvertebrate) A. RICHARD PALMER* Department of Biological Sciences, University of Alberta, Edmonton, AB Canada, T6G 2E9 and Bamfield Marine Station, Bamfield, BC Canada, V0R 1B0 ABSTRACT Phylogenetic analyses of asymmetry varia- lution of lateral bias (DA) must be recognized as distinct from the tion offer a powerful tool for exploring the interplay between evolution of sometimes large but nonetheless random differences ontogeny and evolution because (i) conspicuous asymmetries between sides [antisymmetry (AS)] (7). exist in many higher metazoans with widely varying modes of Genetic and Developmental Aspects. For a trait to evolve, development, (ii) patterns of bilateral variation within species phenotypic variation must be heritable. However, herein lies may identify genetically and environmentally triggered asym- a great puzzle (8). Unlike variation in virtually all other metries, and (iii) asymmetries arising at different times traits, deviations from bilateral symmetry in a particular during development may be more sensitive to internal cyto- direction have not responded to artificial selection. -

First Confirmed Report of a Primary Freshwater Crab (Brachyura: Pseudothelphusidae) Associated with Bromeliads in the Neotropics
JOURNAL OF CRUSTACEAN BIOLOGY, 36(3), 303-309, 2016 FIRST CONFIRMED REPORT OF A PRIMARY FRESHWATER CRAB (BRACHYURA: PSEUDOTHELPHUSIDAE) ASSOCIATED WITH BROMELIADS IN THE NEOTROPICS Ingo S. Wehrtmann 1,2,∗, Célio Magalhães 3, and Orestes Carlos Bello-González 4 1 Museo de Zoología, Escuela de Biología, Universidad de Costa Rica, 11501-2060 San José, Costa Rica 2 Unidad de Investigación Pesquera y Acuicultura (UNIP), Centro de Investigación en Ciencias del Mar y Limnología Downloaded from https://academic.oup.com/jcb/article-abstract/36/3/303/2548966 by guest on 19 May 2020 (CIMAR), Universidad de Costa Rica, 11501-2060 San José, Costa Rica 3 Instituto Nacional de Pesquisas da Amazônia, Caixa Postal 2223, 69080-971 Manaus, Amazonas, Brazil 4 División de Ecología Funcional, Instituto de Ecología y Sistemática, Carretera de Varona, KM 3 1/2, Capdevila, Boyeros, 10 800 La Habana, Cuba ABSTRACT Bromeliads can hold water in their axils, which can harbor an astonishing diversity of macro invertebrates, including decapod crustaceans. Except of a casual report from Trinidad, true freshwater crabs in these phytotelmata have been found exclusively in the Old World, and observations in the New World of crabs associated with bromeliads have been limited to sesarmid crabs, non-primary freshwater crabs that generally do not breed in freshwater. Here we report the presence of a true, or primary, freshwater crab in water-filled axils of a bromeliad in the Neotropics. Immature specimens of the pseudothelphusid crab provisionally identified as Epilobocera cf. gilmanii (Smith, 1870) were collected from inside the bromeliad Hohenbergia penduliflora (A. Richard) Mez. in western Cuba.