An Emerging Pathogen Causing Vesicular Disease in Pigs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Survey of Swine Disease, Management and Biosecurity Practices of Hawai‘I Swine Farms
SURVEY OF SWINE DISEASE, MANAGEMENT AND BIOSECURITY PRACTICES OF HAWAI‘I SWINE FARMS A THESIS SUBMITTED TO THE GRADUATE DIVISION OF THE UNIVERSITY OF HAWAI‘I AT MĀNOA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE IN ANIMAL SCIENCE DECEMBER 2018 By Brittany Amber Castle Thesis Committee: Halina Zaleski, Chairperson Jenee Odani Rajesh Jha Keywords: Swine, Hawai‘i, agriculture ACKNOWLEDGEMENTS I would like to express my deepest gratitude to Dr. Zaleski, my advisor and chair, for her patient guidance and earnest encouragement. Also, thank you for asking for the hard questions which helped me widen my research and thinking. I could not have imagined a better advisor and mentor during my master’s program. I would like to offer my special thanks to the rest of my thesis committee, Dr. Odani and Dr. Jha, for their insightful comments, encouragement, and useful critique of this research. I would like to thank Naomi Ogasawara, the previous graduate student who started this project and who helped lay the groundwork for everything I did. I am particularly grateful for the assistance given to me by Travis Heskett, Laura Ayers, and all the employees of the Hawai‘i Department of Agriculture that provided endless knowledge and my samples for this project. I would like to thank Dr. Fabio Vannucci and the University of Minnesota Veterinary Diagnostic Laboratory their support and sample analysis. Finally, I wish to thank my family and friends for their support, love, and encouragement throughout my study. ii ABSTRACT Although swine diseases and parasites cause significant losses to producers in Hawai‘i, limited information is available on changing disease patterns and related farm practices. -

Seneca Valley Virus
SENECA VALLEY VIRUS Prepared for the Swine Health Information Center By the Center for Food Security and Public Health, College of Veterinary Medicine, Iowa State University DRAFT January, 2016 SUMMARY Etiology • Seneca Valley virus (SVV, also known as Senecavirus A) is a small, non-enveloped picornavirus discovered incidentally in 2002 as a cell culture contaminant. • Only a single species is classified in the genus Senecavirus. The family Picornaviridae also contains foot- and-mouth disease virus (FMDV) and swine vesicular disease virus (SVDV). Cleaning and Disinfection • The efficacy of most disinfectants against SVV is not clearly known. Because vesicular diseases are clinically indistinguishable, disinfection protocols for FMDV should be followed even if SVV is suspected. This includes use of: sodium hydroxide, sodium carbonate, 0.2% citric acid, aldehydes, and oxidizing disinfectants including sodium hypochlorite. • Below are EPA-approved disinfectants USDA lists effective for FMD on page 30 http://www.aphis.usda.gov/animal_health/emergency_management/downloads/fad_epa_disinfectants.pdf. Be sure to follow labeled directions. EPA Reg. No. Product Name Manufacturer Active Ingredient(s) 1677-129 Oxonia Active Ecolab, Inc. Hydrogen peroxide Peroxyacetic acid 6836-86 Lonza DC 101 Lonza, Inc. Alkyl dimethyl benzyl ammonium chloride Didecyl dimethyl ammonium chloride Octyl decyl dimethyl ammonium chloride Dioctyl dimethyl ammonium chloride 10324-67 Maquat MQ615-AS Mason Chemical Company Alkyl dimethyl benzyl ammonium chloride Didecyl dimethyl ammonium chloride Octyl decyl dimethyl ammonium chloride Dioctyl dimethyl ammonium chloride 70060-19 Aseptrol S10-TAB BASF Catalysts, LLC Sodium chlorite Sodium dichloroisocyanurate dehydrate 70060-30 Aseptrol FC-TAB BASF Catalysts, LLC Sodium chlorite Sodium dichloroisocyanurate dehydrate 71654-6 Virkon S E.I. -

) Anguilla Anguilla Isolate from a Diseased European Eel
Characterization of a Novel Picornavirus Isolate from a Diseased European Eel ( Anguilla anguilla) Dieter Fichtner, Anja Philipps, Marco Groth, Heike Schmidt-Posthaus, Harald Granzow, Malte Dauber, Matthias Platzer, Sven M. Bergmann, Daniela Schrudde, Andreas Sauerbrei and Roland Zell J. Virol. 2013, 87(19):10895. DOI: 10.1128/JVI.01094-13. Downloaded from Published Ahead of Print 24 July 2013. Updated information and services can be found at: http://jvi.asm.org/content/87/19/10895 http://jvi.asm.org/ These include: REFERENCES This article cites 47 articles, 20 of which can be accessed free at: http://jvi.asm.org/content/87/19/10895#ref-list-1 CONTENT ALERTS Receive: RSS Feeds, eTOCs, free email alerts (when new on October 28, 2013 by Friedrich-Loeffler-Institut articles cite this article), more» Information about commercial reprint orders: http://journals.asm.org/site/misc/reprints.xhtml To subscribe to to another ASM Journal go to: http://journals.asm.org/site/subscriptions/ Characterization of a Novel Picornavirus Isolate from a Diseased European Eel (Anguilla anguilla) Dieter Fichtner,a Anja Philipps,b* Marco Groth,c Heike Schmidt-Posthaus,d Harald Granzow,a Malte Dauber,e Matthias Platzer,c Sven M. Bergmann,a Daniela Schrudde,a Andreas Sauerbrei,b Roland Zellb Institute of Infectology, Friedrich Loeffler Institut, Federal Research Institute for Animal Health, Greifswald-Insel Riems, Germanya; Department of Virology and Antiviral Therapy, Jena University Hospital, Friedrich Schiller University, Jena, Germanyb; Genome Analysis, Leibniz Institute for Age Research, Fritz Lipmann Institute, Jena, Germanyc; Centre for Fish and Wildlife Health, Institute of Animal Pathology, University of Bern, Bern, Switzerlandd; Institute for Virus Diagnostics, Friedrich Loeffler Institut, Federal Research Institute for Animal Health, Greifswald-Insel Riems, Germanye A novel picornavirus was isolated from specimens of a diseased European eel (Anguilla anguilla). -

Senecavirus A- a Study in Immunogenicity, Seroprevalence, Pathogenesis, and Transmission Elizabeth Rose Houston Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2019 Senecavirus A- a study in immunogenicity, seroprevalence, pathogenesis, and transmission Elizabeth Rose Houston Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Veterinary Medicine Commons, and the Virology Commons Recommended Citation Houston, Elizabeth Rose, "Senecavirus A- a study in immunogenicity, seroprevalence, pathogenesis, and transmission" (2019). Graduate Theses and Dissertations. 17209. https://lib.dr.iastate.edu/etd/17209 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Senecavirus A- a study in immunogenicity, seroprevalence, pathogenesis, and transmission by Elizabeth Houston A thesis submitted to the graduate faculty in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Major: Veterinary Preventive Medicine Program of Study Committee: Pablo Piñeyro, Major Professor James Roth Eric Burrough Luis Giménez-Lirola The student author, whose presentation of the scholarship herein was approved by the program of study committee, is solely responsible for the content of this thesis. The Graduate College will ensure this thesis is globally accessible -

Outbreaks of Neuroinvasive Astrovirus Associated with Encephalomyelitis
Outbreaks of Neuroinvasive Astrovirus Associated with Encephalomyelitis, Weakness, and Paralysis among Weaned Pigs, Hungary Ákos Boros, Mihály Albert, Péter Pankovics, Hunor Bíró, Patricia A. Pesavento, Tung Gia Phan, Eric Delwart, Gábor Reuter A large, highly prolific swine farm in Hungary had a 2-year nervous system (CNS) involvement were reported re- history of neurologic disease among newly weaned (25- to cently in mink, human, bovine, ovine, and swine hosts 35-day-old) pigs, with clinical signs of posterior paraplegia (the latter in certain cases of AII type congenital tremors) and a high mortality rate. Affected pigs that were necropsied (5,6,12–14). Most neuroinvasive astroviruses belong to had encephalomyelitis and neural necrosis. Porcine astrovi- the Virginia/Human-Mink-Ovine (VA/HMO) phyloge- rus type 3 was identified by reverse transcription PCR and in netic clade and cluster with enteric astroviruses identi- situ hybridization in brain and spinal cord samples in 6 ani- mals from this farm. Among tissues tested by quantitative RT- fied from asymptomatic or diarrheic humans and animals PCR, the highest viral loads were detected in brain stem and (15,16). Recent research shows that pigs harbor one of the spinal cord. Similar porcine astrovirus type 3 was also detect- highest astrovirus diversities among mammals examined ed in archived brain and spinal cord samples from another 2 (3,15,20). Porcine astroviruses (PoAstVs) were identified geographically distant farms. Viral RNA was predominantly mainly from diarrheic fecal specimens, less commonly restricted to neurons, particularly in the brain stem, cerebel- from respiratory specimens, although the etiologic role of lum (Purkinje cells), and cervical spinal cord. -
PDF-Document
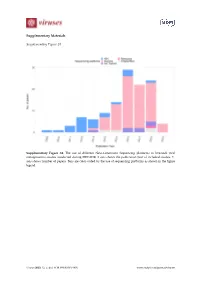
Supplementary Materials Supplementary Figure S1 Supplementary Figure S1. The use of different Next-Generation Sequencing platforms in livestock viral metagenomics studies conducted during 2009-2019. X-axis shows the publication year of included studies. Y- axis shows number of papers. Bars are color-coded by the use of sequencing platforms as shown in the figure legend. Viruses 2020, 12, x; doi: FOR PEER REVIEW www.mdpi.com/journal/viruses Viruses 2020, 12, x FOR PEER REVIEW 2 of 11 Supplementary Figure S2 Small Cattle Poultry Pigs ruminants 100 80 60 Healthy 40 20 0 100 80 GI signs 60 40 20 0 100 80 Respiratory Frequency (%) 60 signs 40 20 0 100 80 clinical signs 60 Other 40 20 0 Specimen types Fecal sample Fetus GI sample Oral sample Respiratory sample Skin sample Blood/plasma/serum/lymph node Genital sample Heart Multiple specimen types Brain Unspecified Supplementary Figure S2. Types of specimens tested in different farm animals by frequency (%), stratified by reported health conditions. Categories per variable are color coded as shown in the legend. GI = gastrointestinal. Viruses 2020, 12, x FOR PEER REVIEW 3 of 11 Search strategies Embase.com (Embase incl. Medline): 1806 ('virome'/de OR 'virus classification'/de OR (('metagenomics'/de OR 'metagenome'/de OR 'high throughput sequencing'/de OR (metagenom* OR ((high-throughput OR shotgun) NEAR/3 (sequenc*))):ab,ti,kw) AND ('virus'/exp OR 'virology'/de OR 'virus infection'/exp OR (virus OR viral* OR virolog*):ab,ti,kw)) OR (virome* OR ((virus* OR viral*) NEAR/3 (classif* OR taxonom* -

A Potential Drug Target for Inhibiting Virus Replication
Old Dominion University ODU Digital Commons Chemistry & Biochemistry Theses & Dissertations Chemistry & Biochemistry Winter 2018 Structure of the Picornavirus Replication Platform: A Potential Drug Target for Inhibiting Virus Replication Meghan Suzanne Warden Old Dominion University, [email protected] Follow this and additional works at: https://digitalcommons.odu.edu/chemistry_etds Part of the Biochemistry Commons, Chemistry Commons, Epidemiology Commons, and the Physiology Commons Recommended Citation Warden, Meghan S.. "Structure of the Picornavirus Replication Platform: A Potential Drug Target for Inhibiting Virus Replication" (2018). Doctor of Philosophy (PhD), Dissertation, Chemistry & Biochemistry, Old Dominion University, DOI: 10.25777/wyvk-8b21 https://digitalcommons.odu.edu/chemistry_etds/22 This Dissertation is brought to you for free and open access by the Chemistry & Biochemistry at ODU Digital Commons. It has been accepted for inclusion in Chemistry & Biochemistry Theses & Dissertations by an authorized administrator of ODU Digital Commons. For more information, please contact [email protected]. STRUCTURE OF THE PICORNAVIRUS REPLICATION PLATFORM: A POTENTIAL DRUG TARGET FOR INHIBITING VIRUS REPLICATION by Meghan Suzanne Warden B.S. May 2011, Lambuth University A Dissertation Submitted to the Faculty of Old Dominion University in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY CHEMISTRY OLD DOMINION UNIVERSITY December 2018 Approved by: Steven M. Pascal (Director) Lesley H. Greene (Member) Hameeda Sultana (Member) James W. Lee (Member) John B. Cooper (Member) ABSTRACT STRUCTURE OF THE PICORNAVIRUS REPLICATION PLATFORM: A POTENTIAL DRUG TARGET FOR INHIBITING VIRUS REPLICATION Meghan Suzanne Warden Old Dominion University, 2018 Director: Dr. Steven M. Pascal Picornaviruses are small, positive-stranded RNA viruses, divided into twelve different genera. -

Efficacy of Three Disinfectants Against Senecavirus a on Five Surfaces and at Two Temperatures
Original research Peer reviewed Efficacy of three disinfectants against Senecavirus A on five surfaces and at two temperatures Azad Singh, MVSc; Sunil K. Mor, PhD; Hamada Aboubakr, MSc; Fabio Vannucci, PhD; Devi P. Patnayak, PhD; Sagar M. Goyal, PhD Summary Results: At ~25°C, household bleach at 1:20 was inactivated within 60 minutes at both Objectives: To evaluate the virucidal effica- dilution inactivated ≥ 99.99% of the virus temperatures and on all surfaces. To detect cy of three commercial disinfectants against within 10 to 15 minutes on aluminum, rub- differences between disinfectants, paired Senecavirus A (SVA) on five different sur- ber, and plastic. On stainless steel and cured Wilcoxon tests were performed. At 10- and faces at ~25°C and 4°C. cement, it inactivated 99.97% and 99.98% 15-minute time points, efficacies of the three of the virus, respectively. At 4°C, bleach disinfectants differed significantly. Materials and methods: Household bleach, inactivated ≥ 99.99% of the virus within a phenolic disinfectant, and a quaternary Implications: Significant variation exists in 5 to15 minutes on all surfaces except rub- ammonium-aldehyde disinfectant were the antiviral efficacies of different disinfec- ber; on rubber, inactivation was 99.91% after tested at manufacturer’s recommended con- tants. Hence, they should be tested against 15 minutes. The phenolic disinfectant at the centrations against a contemporary strain of various pathogens before use in the field. SVA on aluminum, stainless steel, rubber, manufacturer’s recommended concentration cement, and plastic surfaces at ~25°C and inactivated only ≤ 82.41% of the virus at Keywords: swine, Senecavirus A, disinfec- 4°C. -

Wildlife Virology: Emerging Wildlife Viruses of Veterinary and Zoonotic Importance
Wildlife Virology: Emerging Wildlife Viruses of Veterinary and Zoonotic Importance Course #: VME 6195/4906 Class periods: MWF 4:05-4:55 p.m. Class location: Veterinary Academic Building (VAB) Room V3-114 and/or Zoom Academic Term: Spring 2021 Instructor: Andrew Allison, Ph.D. Assistant Professor of Veterinary Virology Department of Comparative, Diagnostic, and Population Medicine College of Veterinary Medicine E-mail: [email protected] Office phone: 352-294-4127 Office location: Veterinary Academic Building V2-151 Office hours: Contact instructor through e-mail to set up an appointment Teaching Assistants: NA Course description: The emergence of viruses that cause disease in animals and humans is a constant threat to veterinary and public health and will continue to be for years to come. The vast majority of recently emerging viruses that have led to explosive outbreaks in humans are naturally maintained in wildlife species, such as influenza A virus (ducks and shorebirds), Ebola virus (bats), Zika virus (non-human primates), and severe acute respiratory syndrome (SARS) coronaviruses (bats). Such epidemics can have severe psychosocial impacts due to widespread morbidity and mortality in humans (and/or domestic animals in the case of epizootics), long-term regional and global economic repercussions costing billions of dollars, in addition to having adverse impacts on vulnerable wildlife populations. Wildlife Virology is a 3-credit (3 hours of lecture/week) undergraduate/graduate-level course focusing on pathogenic viruses that are naturally maintained in wildlife species which are transmissible to humans, domestic animals, and other wildlife/zoological species. In this course, we will cover a comprehensive and diverse set of RNA and DNA viruses that naturally infect free-ranging mammals, birds, reptiles, amphibians, and fish. -

Rna Viral Diversity and Dynamics Along the Antarctic Peninsula
RNA VIRAL DIVERSITY AND DYNAMICS ALONG THE ANTARCTIC PENINSULA A DISSERTATION SUBMITTED TO THE GRADUATE DIVISION OF THE UNIVERSITY OF HAWAIʻI AT MĀNOA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY IN OCEANOGRAPHY MAY 2015 By Jaclyn A. Mueller Dissertation Committee: Grieg Steward, Chairperson Alexander Culley Matthew Church Craig Smith Guylaine Poisson, Outside member Key words: marine RNA viruses, viral ecology, metagenomes, viromes, nucleic acid extraction, reverse transcription quantitative PCR (RT-qPCR), Antarctica © Copyright 2015 – Jaclyn A Mueller All rights reserved. ii ACKNOWLEDGEMENTS This research would not have been possible without the generosity and support of a number of people and organizations, for which I am very thankful. I would first like to acknowledge the Department of Oceanography, the Center for Microbial Oceanography: Research and Education (C-MORE), and the National Science Foundation (NSF) for supporting my research and providing such a fulfilling and enriching graduate career. I am forever indebted to the C-MORE ‘Ohana for their unwavering support and enthusiasm, while providing me the opportunity to experience outreach and education, enhance my leadership and professional development skills, and conduct my doctoral research. In particular, I thank Dr. David Karl for his insights, enthusiasm for oceanography, and continued support of my research. It has been a pleasure to work with so many bright, motivated, and inspiring scientists. I am forever grateful for the patience, guidance, encouragement, and support of my advisor, Dr. Grieg Steward. His enthusiasm and creativity are contagious, and have made this experience quite an enjoyable one. Working with him has not only taught me to be more a perceptive and critical thinker, but also a better writer and scientist. -

N-Linked Glycosylation on Anthrax Toxin Receptor 1 Is Essential for Seneca Valley Virus Infection
viruses Communication N-Linked Glycosylation on Anthrax Toxin Receptor 1 Is Essential for Seneca Valley Virus Infection Nadishka Jayawardena 1,2,†, Linde A. Miles 3,† , Laura N. Burga 1, Charles Rudin 4, Matthias Wolf 2,5,* , John T. Poirier 4,6,* and Mihnea Bostina 1,7,* 1 Department of Microbiology and Immunology, University of Otago, Dunedin 9016, New Zealand; [email protected] (N.J.); [email protected] (L.N.B.) 2 Molecular Cryo-Electron Microscopy Unit, Okinawa Institute of Science and Technology Graduate University, Onna-son, Okinawa 904-0495, Japan 3 Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA; [email protected] 4 Druckenmiller Center for Lung Cancer Research and Department of Medicine, Thoracic Oncology Service, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA; [email protected] 5 Institute of Biological Chemistry, Academia Sinica, Taipei 115, Taiwan 6 Laura and Isaac Perlmutter Cancer Center, New York University Langone Health, New York, NY 10016, USA 7 Otago Micro and Nano Imaging Centre, University of Otago, Dunedin 9016, New Zealand * Correspondence: [email protected] (M.W.); [email protected] (J.T.P.); [email protected] (M.B.) † These authors contributed equally to this work. Abstract: Seneca Valley virus (SVV) is a picornavirus with potency in selectively infecting and lysing cancerous cells. The cellular receptor for SVV mediating the selective tropism for tumors is anthrax toxin receptor 1 (ANTXR1), a type I transmembrane protein expressed in tumors. Similar to Citation: Jayawardena, N.; Miles, other mammalian receptors, ANTXR1 has been shown to harbor N-linked glycosylation sites in its L.A.; Burga, L.N.; Rudin, C.; Wolf, M.; extracellular vWA domain. -

Pdf 10.1371/Journal.Pone.0006883 34
Peer-Reviewed Journal Tracking and Analyzing Disease Trends Pages 1941–2136 EDITOR-IN-CHIEF D. Peter Drotman Associate Editors EDITORIAL BOARD Paul Arguin, Atlanta, Georgia, USA Timothy Barrett, Atlanta, Georgia, USA Charles Ben Beard, Fort Collins, Colorado, USA Barry J. Beaty, Fort Collins, Colorado, USA Ermias Belay, Atlanta, Georgia, USA Martin J. Blaser, New York, New York, USA David Bell, Atlanta, Georgia, USA Richard Bradbury, Atlanta, Georgia, USA Christopher Braden, Atlanta, Georgia, USA Sharon Bloom, Atlanta, GA, USA Arturo Casadevall, New York, New York, USA Mary Brandt, Atlanta, Georgia, USA Kenneth C. Castro, Atlanta, Georgia, USA Corrie Brown, Athens, Georgia, USA Benjamin J. Cowling, Hong Kong, China Charles Calisher, Fort Collins, Colorado, USA Vincent Deubel, Shanghai, China Michel Drancourt, Marseille, France Isaac Chun-Hai Fung, Statesboro, Georgia, USA Paul V. Effler, Perth, Australia Kathleen Gensheimer, College Park, Maryland, USA Anthony Fiore, Atlanta, Georgia, USA Duane J. Gubler, Singapore David Freedman, Birmingham, Alabama, USA Richard L. Guerrant, Charlottesville, Virginia, USA Peter Gerner-Smidt, Atlanta, Georgia, USA Scott Halstead, Arlington, Virginia, USA Stephen Hadler, Atlanta, Georgia, USA Katrina Hedberg, Portland, Oregon, USA Matthew Kuehnert, Atlanta, Georgia, USA David L. Heymann, London, UK Keith Klugman, Seattle, Washington, USA Nina Marano, Atlanta, Georgia, USA Takeshi Kurata, Tokyo, Japan Martin I. Meltzer, Atlanta, Georgia, USA S.K. Lam, Kuala Lumpur, Malaysia David Morens, Bethesda, Maryland, USA Stuart Levy, Boston, Massachusetts, USA J. Glenn Morris, Gainesville, Florida, USA John S. MacKenzie, Perth, Australia Patrice Nordmann, Fribourg, Switzerland John E. McGowan, Jr., Atlanta, Georgia, USA Didier Raoult, Marseille, France Jennifer H. McQuiston, Atlanta, Georgia, USA Pierre Rollin, Atlanta, Georgia, USA Tom Marrie, Halifax, Nova Scotia, Canada Frank Sorvillo, Los Angeles, California, USA Nkuchia M.