Middle Rio Grande Microbial Source Tracking Assessment Report
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

RFP Number and Title: P2013000012, "Biopark Asian Jungle Ropes & Zip Line Course"
` City of Albuquerque Request for Proposals Solicitation Number: P2013000012 BioPark Asian Jungle Ropes & Zip Line Course Deadline for Receipt of Offers October 26, 2012: 4:00 p.m. (Mountain Time) The City eProcurement System will not allow proposals to be submitted after this date and time. Pre-proposal Conference: October 9, 2012 2:30 PM Albuquerque BioPark 903 Tenth Street Albuquerque, NM 87102 Library City of Albuquerque P2013000012, " BioPark Asian Jungle Ropes & Zip Line Course” 1 Department of Finance and Administrative Services Purchasing Division TABLE OF CONTENTS Page Introduction 3 Part 1 Instructions to Offerors 4 Part 2 Proposal Format 13 Part 3 Scope of Services 15 Part 4 Evaluation of Offers 16 Part 5 Local Preference Instructions 18 Local Preference Form 19 Part 6 Draft Agreement 22 Appendix A Revenue Proposal Forms 28 Appendix B Bond Forms 29 P2013000012, " BioPark Asian Jungle Ropes & Zip Line Course” 2 INTRODUCTION Purpose and Intent The Albuquerque Biological Park (BioPark) is soliciting proposals from qualified firms interested in providing an Asian Jungle Experience using a combination of rope courses and zip lines on a 2 acre parcel of land located at the Rio Grande Zoo, 903 10th St. Albuquerque NM 87102. The Albuquerque Biological Park is an accredited facility through the Association of Zoos and Aquariums (AZA) and seeks to maintain and continually improve upon the care and management of the animal and plant collection as well as providing an important recreational and educational venue to the citizens and visitors of the Albuquerque region. Background and General Information 1. The Albuquerque Biological Park is owned and operated by the City of Albuquerque. -

General Vertical Files Anderson Reading Room Center for Southwest Research Zimmerman Library
“A” – biographical Abiquiu, NM GUIDE TO THE GENERAL VERTICAL FILES ANDERSON READING ROOM CENTER FOR SOUTHWEST RESEARCH ZIMMERMAN LIBRARY (See UNM Archives Vertical Files http://rmoa.unm.edu/docviewer.php?docId=nmuunmverticalfiles.xml) FOLDER HEADINGS “A” – biographical Alpha folders contain clippings about various misc. individuals, artists, writers, etc, whose names begin with “A.” Alpha folders exist for most letters of the alphabet. Abbey, Edward – author Abeita, Jim – artist – Navajo Abell, Bertha M. – first Anglo born near Albuquerque Abeyta / Abeita – biographical information of people with this surname Abeyta, Tony – painter - Navajo Abiquiu, NM – General – Catholic – Christ in the Desert Monastery – Dam and Reservoir Abo Pass - history. See also Salinas National Monument Abousleman – biographical information of people with this surname Afghanistan War – NM – See also Iraq War Abousleman – biographical information of people with this surname Abrams, Jonathan – art collector Abreu, Margaret Silva – author: Hispanic, folklore, foods Abruzzo, Ben – balloonist. See also Ballooning, Albuquerque Balloon Fiesta Acequias – ditches (canoas, ground wáter, surface wáter, puming, water rights (See also Land Grants; Rio Grande Valley; Water; and Santa Fe - Acequia Madre) Acequias – Albuquerque, map 2005-2006 – ditch system in city Acequias – Colorado (San Luis) Ackerman, Mae N. – Masonic leader Acoma Pueblo - Sky City. See also Indian gaming. See also Pueblos – General; and Onate, Juan de Acuff, Mark – newspaper editor – NM Independent and -

Publication of the Utica Zoological Society Volume 4 Number 3
Publication of the Utica Zoological Society May/June 1996 Volume 4 Number 3 ~..,.- Spring has arrived ...~nally! It~~--~,._ sensitive. The babies have tender was a long, cold wmter, bu -::: ~-. v~ ~ . ink skin and are easily sunburned underneath several ~eet of snow, · - ~· - , e;' ' uring their first few weeks of life. plants and arumals are To prevent this mom will lead her beginning to feel the change of the whale . family, are usually baby to a mud hoie so they can cover seasons.For many species of wildlife, h~lthy and robust ~t birth. Th~y themselves with~ pr?,tective co~ting of spring is the time for giving birth. The qm~kly adapt t? their new aq~IC mud, sort of a rhino sunscreen . ycle of life is quite different for envrronment, With mother watching When they are born, African • mammals, birds, fish, amphibians and them very closely. The mother colobus monkeys are covered with reptiles. All mammals (with the guides her calfs movements, and curly white fur! The infants are passed exception of a few oddities such as the some mothers have even been back and forth among the troop's duck-billed platypus and the spiny observed disciplining the calves females, and it is believed that the all- ~teater) give birth to l.iv~ young, while when they misbehave! The calf white fur of the babies encourages this birds, fish and amp~tbians lay eg~s . nurses within an hour after it is behavior. Because they look different Snakes are an exception: some spectes . · b'rth t . born, and as often as four times from the adults, who are black With gtve 1 o 1IVe young, some 1ay eggs, . -

Environmental Contaminants and Their Effects on Fish in the Rio Grande Basin
Biomonitoring of Environmental Status and Trends (BEST) Program: Environmental Contaminants and their Effects on Fish in the Rio Grande Basin S# S# S# S# S#S#S# S#S#S#S# S# S# # S S# S# # S S# S# S# S# # S# S# S S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# S# # S# S# # S S#S# S S# S# S# S# #S# S# S# S# S#S S# # S# SS# S# S# S#S# Scientific Investigations Report 2004—5108 U.S. Department of the Interior U.S. Geological Survey Front cover. The U.S. map shows the Rio Grande Basin (green) and stations sampled in this study (orange). Shown in gray are major river basins and stations in the conterminous U.S. sampled during other Biomonitoring of Environmental Status and Trends Program (BEST) investigations. Biomonitoring of Environmental Status and Trends (BEST) Program: Environmental Contaminants and their Effects on Fish in the Rio Grande Basin By Christopher J. Schmitt, Gail M. Dethloff, Jo Ellen Hinck, Timothy M. Bartish, Vicki S. Blazer, James J. Coyle, Nancy D. Denslow, and Donald E. Tillitt Scientific Investigations Report 2004—5108 U.S. Department of the Interior U.S. Geological Survey U.S. Department of the Interior Gale A. Norton, Secretary U.S. Geological Survey Charles G. Groat, Director U.S. Geological Survey, Reston, Virginia: 2004 For more information about the USGS and its products: Telephone: 1-888-ASK-USGS World Wide Web: http://www.usgs.gov/ Any use of trade, product, or firm names in this publication is for descriptive purposes only and does not imply endorsement by the U.S. -
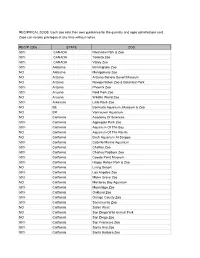
2006 Reciprocal List
RECIPRICAL ZOOS. Each zoo sets their own guidelines for the quantity and ages admitted per card. Zoos can revoke privileges at any time without notice. RECIP 2006 STATE ZOO 50% CANADA Riverview Park & Zoo 50% CANADA Toronto Zoo 50% CANADA Valley Zoo 50% Alabama Birmingham Zoo NO Alabama Montgomery Zoo NO Arizona Arizona-Sonora Desert Museum NO Arizona Navajo Nation Zoo & Botanical Park 50% Arizona Phoenix Zoo 50% Arizona Reid Park Zoo NO Arizona Wildlife World Zoo 50% Arkansas Little Rock Zoo NO BE Bermuda Aquarium, Museum & Zoo NO BR Vancouver Aquarium NO California Academy Of Sciences 50% California Applegate Park Zoo 50% California Aquarium Of The Bay NO California Aquarium Of The Pacific NO California Birch Aquarium At Scripps 50% California Cabrillo Marine Aquarium 50% California Chaffee Zoo 50% California Charles Paddock Zoo 50% California Coyote Point Museum 50% California Happy Hollow Park & Zoo NO California Living Desert 50% California Los Angeles Zoo 50% California Micke Grove Zoo NO California Monterey Bay Aquarium 50% California Moonridge Zoo 50% California Oakland Zoo 50% California Orange County Zoo 50% California Sacramento Zoo NO California Safari West NO California San Diego Wild Animal Park NO California San Diego Zoo 50% California San Francisco Zoo 50% California Santa Ana Zoo 50% California Santa Barbara Zoo NO California Seaworld San Diego 50% California Sequoia Park Zoo NO California Six Flags Marine World NO California Steinhart Aquarium NO CANADA Calgary Zoo 50% Colorado Butterfly Pavilion NO Colorado Cheyenne -

Kirtland Inn Guestbook
Dear Guest, Welcome to the Kirtland Inn, setting the standard for the USAF! On behalf of the staff, we sincerely hope your stay is pleasant and comfortable. Kirtland Inn and Kirtland Air Force Base are rich in history and tradition. Please take time out of your busy schedule to enjoy the sights and the surrounding area. We strive to provide outstanding guest service and accommodations to all of our patrons. Should you need anything special or if you have inadvertently forgotten a travel item, please ask any staff member for assistance. We will take care of your request as quickly as possible. If you need further information about Kirtland Air Force Base and the surrounding community, just ask. Continuous improvement is our goal. We welcome any comments or recommendations that you may have. Please complete the comment card located in your room or at the Reception Desks to let us know how we did. You can reach me at 846-9663 or any of our Guest Service Representatives by dialing 0. Again, welcome to Kirtland AFB and the Kirtland Inn! Steve Holland, CHA General Manager Email: [email protected] The appearance of local business names does NOT imply federal endorsements. All information to include addresses and telephone numbers are subject to change. Please call the business to confirm their operation hours. GENERAL INFORMATION This information is made available as a public service and does not imply Air Force endorsement of the company’s products or services. CALL 853-9111 FOR ANY ON BASE EMERCENCY Kirtland AFB Gate Information Gate Phone Eubank Gate 846-6231 Gibson Gate 846-7240 Maxwell Gate 846-7491 Truman Gate 846-7509 Wyoming Gate 846-6118 Receiving Mail During Your Stay Guests who require mail service while staying at the Kirtland Inn should use the base general delivery address: Your Name General Delivery 2050 2nd St SE Kirtland Air Force Base, NM 87117 *Upon arrival to Kirtland AFB, individuals utilizing general delivery mail service must come by the Postal Service Center, building 20204, to fill out a locator card with your new address. -

1 Written Testimony Submitted to the United States Senate Committee On
Written Testimony Submitted to the United States Senate Committee on Energy and Natural Resources on S. 1012 New Mexico Drought Preparedness Act of 2017 Respectfully Submitted By Mike A. Hamman, PE Chief Executive Officer Middle Rio Grande Conservancy District Rio Grande Water Development in New Mexico The Upper Rio Grande originates in the San Juan and Sangre de Cristo mountain ranges in southern Colorado and northern New Mexico. It bisects the San Luis Valley in Colorado and the entire state of New Mexico with this reach culminating at Fort Quitman, Texas. This portion of the Rio Grande is administered under the Rio Grande Compact by a federal appointee and three Commissioners from Colorado, New Mexico and Texas with support from the United States Geological Survey, the Bureau of Reclamation, and the Army Corps of Engineers. The annual mean flow as measured at the Otowi gage in New Mexico is 1 million acre-feet with wide variation, ranging from 250,000 to 2.5 million acre-feet. Irrigated agriculture consists of approximately 600,000 acres in Colorado, 200,000 acres in New Mexico, 100,000 acres in Texas. Additionally, up to 60,000 acre-feet is delivered to lands within the Republic of Mexico via the Rio Grande Project under the 1906 Convention between the United States and Mexico. The predominate crop due to climate, water supplies and labor considerations is alfalfa. Other crops include potatoes, chilé, corn, fruit, onions and pecans. There is an improving ‘farm to table’ market serving a demand for locally produced agricultural products ranging from lettuces to melons as well as organically grown products particularly near and in municipalities. -

2002 Federal Register, 67 FR 39205; Centralized Library: U.S. Fish
Thursday, June 6, 2002 Part IV Department of the Interior Fish and Wildlife Service 50 CFR Part 17 Endangered and Threatened Wildlife and Plants; Designation of Critical Habitat for the Rio Grande Silvery Minnow; Proposed Rule VerDate May<23>2002 18:22 Jun 05, 2002 Jkt 197001 PO 00000 Frm 00001 Fmt 4717 Sfmt 4717 E:\FR\FM\06JNP3.SGM pfrm17 PsN: 06JNP3 39206 Federal Register / Vol. 67, No. 109 / Thursday, June 6, 2002 / Proposed Rules DEPARTMENT OF THE INTERIOR ADDRESSES: 1. Send your comments on fishes in the Rio Grande Basin, this proposed rule, the draft economic occurring from Espan˜ ola, NM, to the Fish and Wildlife Service analysis, and draft EIS to the New Gulf of Mexico (Bestgen and Platania Mexico Ecological Services Field Office, 1991). It was also found in the Pecos 50 CFR Part 17 2105 Osuna Road NE, Albuquerque, River, a major tributary of the Rio RIN 1018–AH91 NM, 87113. Written comments may also Grande, from Santa Rosa, NM, be sent by facsimile to (505) 346–2542 downstream to its confluence with the Endangered and Threatened Wildlife or through the Internet to Rio Grande (Pflieger 1980). The silvery and Plants; Designation of Critical [email protected]. You may also minnow is completely extirpated from Habitat for the Rio Grande Silvery hand-deliver written comments to our the Pecos River and from the Rio Grande Minnow New Mexico Ecological Services Field downstream of Elephant Butte Reservoir Office, at the above address. You may and upstream of Cochiti Reservoir AGENCY: Fish and Wildlife Service, obtain copies of the proposed rule, the (Bestgen and Platania 1991). -

Rio Grande Heritage Farm Education Guide
Rio Grande Heritage Farm Education Guide Grades 4-5 Contents General Biopark Information Biopark Group Admission Information School Lunch Order Form General Teacher Tips Chaperone Guide Teacher Background Information Pre-Visit Activities Field Trip Worksheet Post-Visit Activities Student Handouts Student Handout Answer Sheets GENERAL BIOPARK INFORMATION Facilities The Albuquerque BioPark is a gateway to the mystery and diversity of living organisms. Core facilities include: Albuquerque Aquarium, Rio Grande Botanic Garden, Rio Grande Zoo and Tingley Beach. The BioPark has an estimated 6,000 animals, 11,000 plants, 300 staff and 300 year-round volunteers. The Director of the Albuquerque BioPark is Ray Darnell. The Albuquerque Aquarium is a great place to explore aquatic environments by tracing a drop of water from the headwaters of the Rio Grande in southern Colorado to the Gulf of Mexico and out to the oceans of the world. Current exhibits highlight fish of the Rio Grande at Central Bridge, moray eels, invertebrates, coral reefs, floating jellies, sea turtles and lots of toothy sharks. Further construction will develop tanks that highlight species that live in the Pacific Ocean. (Location 2601 Central NW) The Rio Grande Botanic Garden celebrates the miracle of photosynthesis in living color every day. Our state-of-the-art glass conservatories feature plants native to Mediterranean climate zones and xeric plants from North American deserts and other arid regions of the world. A trio of formal walled gardens illustrates Old World design in fountains, tile, herbs and roses. El Jardin de la Curandera is an ethnobotanic exhibit with a beautiful bronze sculpture, and the Children’s Fantasy Garden has giant fun for kids of all ages. -

Calendar Year 2016 Report to the Rio Grande Compact Commission
University of New Mexico UNM Digital Repository Law of the Rio Chama The Utton Transboundary Resources Center 2016 Calendar Year 2016 Report to the Rio Grande Compact Commission Dick Wolfe Colorado Tom Blaine New Mexico Patrick R. Gordon Texas Hal Simpson Federal Chairman Follow this and additional works at: https://digitalrepository.unm.edu/uc_rio_chama Recommended Citation Wolfe, Dick; Tom Blaine; Patrick R. Gordon; and Hal Simpson. "Calendar Year 2016 Report to the Rio Grande Compact Commission." (2016). https://digitalrepository.unm.edu/uc_rio_chama/72 This Report is brought to you for free and open access by the The Utton Transboundary Resources Center at UNM Digital Repository. It has been accepted for inclusion in Law of the Rio Chama by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected], [email protected], [email protected]. Calendar Year 2016 Report to the Rio Grande Compact Commission Colorado New Mexico Texas Dick Wolfe Tom Blaine Patrick R. Gordon Federal Chairman Hal Simpson U. S. Department of the Interior Bureau of Reclamation Albuquerque Area Office Albuquerque, New Mexico March 2017 MISSION STATEMENTS The Department of the Interior protects and manages the Nation's natural resources and cultural heritage; provides scientific and other information about those resources; and honors its trust responsibilities or special commitments to American Indians, Alaska Natives, and affiliated island communities. The mission of the Bureau of Reclamation is to manage, develop, and protect water and related resources in an environmentally and economically sound manner in the interest of the American public. Cover photo – Overbank flow at habitat restoration site on the Sevilleta NWR during 2016 spring pulse (Dustin Armstrong, Reclamation) Calendar Year 2016 Report to the Rio Grande Compact Commission U. -

Numerical Modeling of Isleta Diversion Dam Gate Operation Hydraulics to Minimize Sediment Effects
NUMERICAL MODELING OF ISLETA DIVERSION DAM GATE OPERATION HYDRAULICS TO MINIMIZE SEDIMENT EFFECTS Drew C. Baird, Hydraulic Engineer, U.S. Bureau of Reclamation, Denver, Colorado, [email protected]; Michael Sixta, Hydraulic Engineer, U.S. Bureau of Reclamation, Denver, Colorado, [email protected]. INTRODUCTION Isleta Diversion Dam was constructed in 1934 by the Middle Rio Grande Conservancy District (MRGCD) as part of their irrigation system, and is located on the Rio Grande about 10 miles south of Albuquerque, New Mexico, immediately downstream from the Highway 147 Bridge (Figure 1). The diversion dam was rehabilitated by Reclamation in 1955 as part of the Middle Rio Grande Project, authorized by Congress in the 1948 and 1950 Flood Control Acts. The Middle Rio Grande (MRG) has long been recognized for its characteristics of high sediment loads and dynamic channel conditions (Happ, 1948; Lagasse, 1980; Makar, 2010). The Isleta Diversion Dam consists of 30 river gates, three headworks gates on the Peralta Main canal (east side), and four headworks gates on the Belen Highline canal (west side) of the dam (Figure 2). The headworks gates are located in a sluiceway with a downstream gate used to maintain a maximum diversion head. Gate operations are used to provide water to downstream irrigators, meet downstream flow requirements of the 2003 Endangered Species Biological Opinion (USFWS, 2003), and manage sediment. Within the context of these multiple water use needs, a one-dimensional (1D) and two-dimensional (2D) fixed bed hydraulic models, sluiceway hydraulics, and sediment incipient motion analysis has been completed to provide recommendations on gate operations that would help reduce sediment impacts. -

Water Resources of the Middle Rio Grande 38 Chapter Two
THE MIDDLE RIO GRANDE TODAY 37 Infrastructure and Management of the Middle Rio Grande Leann Towne, U.S. Bureau of Reclamation any entities are involved in water management lands within the Middle Rio Grande valley from M in the Middle Rio Grande valley from Cochiti to Cochiti Dam to the Bosque del Apache National Elephant Butte Reservoir. These entities own and Wildlife Refuge. The four divisions are served by operate various infrastructure in the Middle Rio Middle Rio Grande Project facilities, which consist of Grande valley that are highly interconnected and ulti- the floodway and three diversion dams, more than mately affect water management of the Rio Grande. 780 miles of canals and laterals, and almost 400 miles This paper describes major hydrologic aspects of the of drains. Users are served by direct diversions from Middle Rio Grande valley, including water manage- the Rio Grande and from internal project flows such ment activities of the U.S. Bureau of Reclamation, as drain returns. These irrigation facilities are operated major infrastructure of the Middle Rio Grande Project and maintained by MRGCD. (including the Low Flow Conveyance Channel), and focusing on issues downstream of San Acacia COCHITI DIVISION Diversion Dam. Although other entities such as municipalities have significant water management Project diversions from the Rio Grande begin at responsibilities in the Middle Rio Grande valley, they Cochiti Dam, through two canal headings that serve will not be addressed in this paper. the Cochiti Division. The Cochiti East Side Main and The Middle Rio Grande Conservancy District, a Sile Main canals deliver water to irrigators on both political subdivision of the state of New Mexico, was sides of the Rio Grande.