Nodal Protein Processing and Fibroblast Growth Factor 4 Synergize to Maintain a Trophoblast Stem Cell Microenvironment
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Biology of Hepatocellular Carcinoma: Implications for Genomic and Immune Therapies Galina Khemlina1,4*, Sadakatsu Ikeda2,3 and Razelle Kurzrock2
Khemlina et al. Molecular Cancer (2017) 16:149 DOI 10.1186/s12943-017-0712-x REVIEW Open Access The biology of Hepatocellular carcinoma: implications for genomic and immune therapies Galina Khemlina1,4*, Sadakatsu Ikeda2,3 and Razelle Kurzrock2 Abstract Hepatocellular carcinoma (HCC), the most common type of primary liver cancer, is a leading cause of cancer-related death worldwide. It is highly refractory to most systemic therapies. Recently, significant progress has been made in uncovering genomic alterations in HCC, including potentially targetable aberrations. The most common molecular anomalies in this malignancy are mutations in the TERT promoter, TP53, CTNNB1, AXIN1, ARID1A, CDKN2A and CCND1 genes. PTEN loss at the protein level is also frequent. Genomic portfolios stratify by risk factors as follows: (i) CTNNB1 with alcoholic cirrhosis; and (ii) TP53 with hepatitis B virus-induced cirrhosis. Activating mutations in CTNNB1 and inactivating mutations in AXIN1 both activate WNT signaling. Alterations in this pathway, as well as in TP53 and the cell cycle machinery, and in the PI3K/Akt/mTor axis (the latter activated in the presence of PTEN loss), as well as aberrant angiogenesis and epigenetic anomalies, appear to be major events in HCC. Many of these abnormalities may be pharmacologically tractable. Immunotherapy with checkpoint inhibitors is also emerging as an important treatment option. Indeed, 82% of patients express PD-L1 (immunohistochemistry) and response rates to anti-PD-1 treatment are about 19%, and include about 5% complete remissions as well as durable benefit in some patients. Biomarker-matched trials are still limited in this disease, and many of the genomic alterations in HCC remain challenging to target. -

The Roles of Fgfs in the Early Development of Vertebrate Limbs
Downloaded from genesdev.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW The roles of FGFs in the early development of vertebrate limbs Gail R. Martin1 Department of Anatomy and Program in Developmental Biology, School of Medicine, University of California at San Francisco, San Francisco, California 94143–0452 USA ‘‘Fibroblast growth factor’’ (FGF) was first identified 25 tion of two closely related proteins—acidic FGF and ba- years ago as a mitogenic activity in pituitary extracts sic FGF (now designated FGF1 and FGF2, respectively). (Armelin 1973; Gospodarowicz 1974). This modest ob- With the advent of gene isolation techniques it became servation subsequently led to the identification of a large apparent that the Fgf1 and Fgf2 genes are members of a family of proteins that affect cell proliferation, differen- large family, now known to be comprised of at least 17 tiation, survival, and motility (for review, see Basilico genes, Fgf1–Fgf17, in mammals (see Coulier et al. 1997; and Moscatelli 1992; Baird 1994). Recently, evidence has McWhirter et al. 1997; Hoshikawa et al. 1998; Miyake been accumulating that specific members of the FGF 1998). At least five of these genes are expressed in the family function as key intercellular signaling molecules developing limb (see Table 1). The proteins encoded by in embryogenesis (for review, see Goldfarb 1996). Indeed, the 17 different FGF genes range from 155 to 268 amino it may be no exaggeration to say that, in conjunction acid residues in length, and each contains a conserved with the members of a small number of other signaling ‘‘core’’ sequence of ∼120 amino acids that confers a com- molecule families [including WNT (Parr and McMahon mon tertiary structure and the ability to bind heparin or 1994), Hedgehog (HH) (Hammerschmidt et al. -

And Fibroblast Growth Factor Receptor Interactions Usin
www.nature.com/scientificreports OPEN Characterization of clostridium botulinum neurotoxin serotype A (BoNT/A) and fbroblast growth factor receptor interactions using novel receptor dimerization assay Nicholas G. James1, Shiazah Malik2, Bethany J. Sanstrum1, Catherine Rhéaume2, Ron S. Broide2, David M. Jameson1, Amy Brideau‑Andersen2 & Birgitte S. Jacky2* Clostridium botulinum neurotoxin serotype A (BoNT/A) is a potent neurotoxin that serves as an efective therapeutic for several neuromuscular disorders via induction of temporary muscular paralysis. Specifc binding and internalization of BoNT/A into neuronal cells is mediated by its binding domain (HC/A), which binds to gangliosides, including GT1b, and protein cell surface receptors, including SV2. Previously, recombinant HC/A was also shown to bind to FGFR3. As FGFR dimerization is an indirect measure of ligand‑receptor binding, an FCS & TIRF receptor dimerization assay was developed to measure rHC/A‑induced dimerization of fuorescently tagged FGFR subtypes (FGFR1‑ 3) in cells. rHC/A dimerized FGFR subtypes in the rank order FGFR3c (EC50 ≈ 27 nM) > FGFR2b (EC50 ≈ 70 nM) > FGFR1c (EC50 ≈ 163 nM); rHC/A dimerized FGFR3c with similar potency as the native FGFR3c ligand, FGF9 (EC50 ≈ 18 nM). Mutating the ganglioside binding site in HC/A, or removal of GT1b from the media, resulted in decreased dimerization. Interestingly, reduced dimerization was also observed with an SV2 mutant variant of HC/A. Overall, the results suggest that the FCS & TIRF receptor dimerization assay can assess FGFR dimerization with known and novel ligands and support a model wherein HC/A, either directly or indirectly, interacts with FGFRs and induces receptor dimerization. Botulinum neurotoxin type A (BoNT/A) is a 150 kDa metalloenzyme belonging to the family of neurotoxins produced by Clostridium botulinum. -

In Vitro Differentiation of Human Umbilical Cord Blood Mesenchymal
Alexandria Journal of Medicine (2017) 53, 167–173 HOSTED BY Alexandria University Faculty of Medicine Alexandria Journal of Medicine http://www.elsevier.com/locate/ajme In vitro differentiation of human umbilical cord blood mesenchymal stem cells into functioning hepatocytes May H. Hasan a, Kamal G. Botros a, Mona A. El-Shahat a,*, Hussein A. Abdallah b, Mohamed A. Sobh c a Anatomy and Embryology Department, Faculty of Medicine, Mansoura University, Egypt b Medical Biochemistry Department, Faculty of Medicine, Mansoura University, Egypt c Experimental Biology, Urology & Nephrology Center, Mansoura University, Egypt Received 29 January 2016; revised 6 May 2016; accepted 14 May 2016 Available online 5 August 2016 KEYWORDS Abstract Mesenchymal stem cells (MSCs) were isolated by gradient density centrifugation from Umbilical cord blood; umbilical cord blood. Spindle-shaped adherent cells were permitted to grow to 70% confluence Mesenchymal stem cells; in primary culture media which was reached by day 12. Induction of differentiation started by cul- Culture; turing cells with differentiation medium containing FGF-4 and HGF. Under hepatogenic condi- Hepatocytes; tions few cuboidal cells appeared in culture on day 7. From day 21 to day 28, most of cells HGF; became small and round. The control negative cells cultured in serum free media showed FGF-4 fibroblast-like morphology. Urea production and protein secretion by the differentiated hepatocyte-like cells were detected on day 21 and increased on day 28. Protein was significantly increased in comparison with control by day 28. The cells became positive for AFP at day 7 and positive cells could still be detected at days 21 and 28. -

Investigation of the Role of Capicua in the FGF Signalling Pathway and the Wound Response
Investigation of the role of Capicua in the FGF signalling pathway and the wound response Laura May-Seen Cowell Master by Research University of York Biology September 2019 Abstract Fibroblast growth factor (FGF) signalling is critical for the initiation and regulation of multiple developmental processes including gastrulation, mesoderm induction and limb development. Despite extensive understanding of FGF signal transduction via tyrosine kinase receptors (RTKs), the specific mechanism responsible for regulation of target gene transcription is still not fully understood. The protein Capicua (CIC) has been linked to transcriptional regulation in RTK signalling via the ERK pathway in multiple organisms. ERK signalling cascades also mediate wound signal transduction and transcription of associated genes. We hypothesise that transcription of a subset of FGF target genes, and genes involved in the wound response, rely on ERK mediated relief of CIC transcriptional repression. The aims of this project were to establish if CIC operates downstream of FGF signalling through analysis and validation of RNA-seq data, and to investigate the relationship between CIC and ERK in FGF signalling and wound healing in Xenopus embryos. The work in this thesis shows that CIC knockdown and FGF overexpressing embryos exhibit similar phenotypes and transcriptomes. Immunostaining for myc-tagged CIC indicates that CIC expression is reduced following activation of FGF signalling or the wound response. 75% of the putative CIC and FGF regulated genes analysed have enriched CIC binding sites and 75% were upregulated in RT- PCR analysis of CIC knockdown and FGF overexpressing embryos. Additionally, genes associated with wound healing (fos and gadd45a) are upregulated in CIC knockdown embryos. -

Post-Transcriptional Tuning of FGF Signaling Mediates Neural Crest Induction
Post-transcriptional tuning of FGF signaling mediates neural crest induction Jacqueline Copelanda and Marcos Simoes-Costaa,1 aDepartment of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14850 Edited by Janet Rossant, The Gairdner Foundation, Toronto, Canada, and approved November 16, 2020 (received for review May 20, 2020) Ectodermal patterning is required for the establishment of multi- In recent years, growing evidence has supported a role for ple components of the vertebrate body plan. Previous studies have microRNA (miRNA)-mediated gene silencing in neural crest demonstrated that precise combinations of extracellular signals development (18–21). In the developing mouse embryo, the induce distinct ectodermal cell populations, such as the neural tissue-specific deletion of DICER, a key enzyme in miRNA crest and the neural plate. Yet, we still lack understanding of how biogenesis, results in a complete loss or malformation of several the response to inductive signals is modulated to generate the neural crest-derived structures (22–25). Furthermore, several proper transcriptional output in target cells. Here we show that lines of evidence have underscored a decisive role for specific posttranscriptional attenuation of fibroblast growth factor (FGF) miRNAs in modulating neural crest stem cell identity (19, 21, signaling is essential for the establishment of the neural crest 26). In other developmental contexts, miRNAs have been shown territory. We found that neural crest progenitors display elevated to modulate the expression of components of signaling cascades, expression of DICER, which promotes enhanced maturation of a thus influencing developmental decisions (27–30). These results set of cell-type-specific miRNAs. These miRNAs collectively target raise the possibility that posttranscriptional mechanisms may components of the FGF signaling pathway, a central player in the play an important role in the formation of ectodermal deriva- process of neural induction in amniotes. -
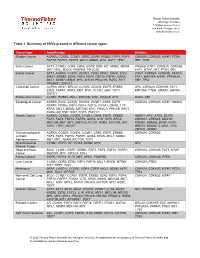
Table 1: Summary of Cnvs Present in Different Cancer Types
Thermo Fisher Scientific Oncology Informatics 110 Miller Avenue, Floor 2 Ann Arbor, Michigan 48104 www.thermofisher.com Table 1: Summary of CNVs present in different cancer types Cancer type Amplification Deletion Bladder Cancer AURKA, CCND1, CCNE1, DDR2, EGFR, ERBB2, FGF3, FGF4, CDKN2A, CDKN2B, MTAP, PTEN, FGF19, FGFR1, FGFR3, MCL1, MDM2, MYC, RAF1, TERT RB1, TP53 Brain Cancer AKT3, CCND2, CDK4, CDK6, EGFR, KDR, KIT, MDM2, MDM4, ARID5B, ATRX, CDKN2A, CDKN2B, MET, MYC, MYCN, PDGFRA, PIK3C2B FAT1, MTAP, NF1, PTEN, RB1 Breast Cancer AKT3, AURKA, CCND1, CCNE1, CDK4, DDR2, EGFR, ELF3, CDH1, CDKN2A, CDKN2B, FANCA, EMSY, ERBB2, EZH2, FGF3, FGF4, FGF19, FGFR1, GNAS, FAT1, MAP2K4, MTAP, PPP2R2A, MCL1, MDM2, MDM4, MYC, MYCN, PIK3C2B, RARA, RIT1, RB1, TP53 RPS6KB1, ZNF217 Colorectal Cancer AURKA, BRAF, BRCA2, CCND2, CCND3, EGFR, ERBB2, APC, CDKN2A, CDKN2B, FAT1, EZH2, FGFR1, GNAS, MET, MYC, PLCG1, SRC, TOP1, MAP2K4, PTEN, SMAD2, SMAD4 ZNF217 Endometrial Cancer CCNE1, ERBB2, MCL1, MECOM, MYC, PIK3CB, RIT1 DAXX, RB1 Esophageal Cancer AURKA, BCL6, CCND1, CCND3, CCNE1, CDK6, EGFR, CDKN2A, CDKN2B, MTAP, SMAD4 ERBB2, ERBB3, FGF3, FGF4, FGF19, FOXA1, GNAS, IL7R, KRAS, MCL1, MDM2, MECOM, MYC, PIK3CA, PIK3CB, RAC1, RARA, RICTOR, TERT, TOP1, ZNF217 Gastric Cancer AURKA, CCND1, CCND3, CCNE1, CDK6, EGFR, ERBB2, AMER1, APC, ATRX, BCOR, FGF3, FGF4, FGF19, FGFR2, GNAS, IL7R, KLF5, KRAS, CDKN2A, CDKN2B, DDX3X, MECOM, MET, MYC, PIK3CA, PLCG1, RARA, RICTOR, SRC, KDM5C, KDM6A, MTAP, PHF6, TERT, TOP1, ZNF217 RBM10, SMAD4, STAG2, TP53, ZMYM3, ZRSR2 Gastroesophageal -

FGF/FGFR Signaling in Health and Disease
Signal Transduction and Targeted Therapy www.nature.com/sigtrans REVIEW ARTICLE OPEN FGF/FGFR signaling in health and disease Yangli Xie1, Nan Su1, Jing Yang1, Qiaoyan Tan1, Shuo Huang 1, Min Jin1, Zhenhong Ni1, Bin Zhang1, Dali Zhang1, Fengtao Luo1, Hangang Chen1, Xianding Sun1, Jian Q. Feng2, Huabing Qi1 and Lin Chen 1 Growing evidences suggest that the fibroblast growth factor/FGF receptor (FGF/FGFR) signaling has crucial roles in a multitude of processes during embryonic development and adult homeostasis by regulating cellular lineage commitment, differentiation, proliferation, and apoptosis of various types of cells. In this review, we provide a comprehensive overview of the current understanding of FGF signaling and its roles in organ development, injury repair, and the pathophysiology of spectrum of diseases, which is a consequence of FGF signaling dysregulation, including cancers and chronic kidney disease (CKD). In this context, the agonists and antagonists for FGF-FGFRs might have therapeutic benefits in multiple systems. Signal Transduction and Targeted Therapy (2020) 5:181; https://doi.org/10.1038/s41392-020-00222-7 INTRODUCTION OF THE FGF/FGFR SIGNALING The binding of FGFs to the inactive monomeric FGFRs will Fibroblast growth factors (FGFs) are broad-spectrum mitogens and trigger the conformational changes of FGFRs, resulting in 1234567890();,: regulate a wide range of cellular functions, including migration, dimerization and activation of the cytosolic tyrosine kinases by proliferation, differentiation, and survival. It is well documented phosphorylating the tyrosine residues within the cytosolic tail of that FGF signaling plays essential roles in development, metabo- FGFRs.4 Then, the phosphorylated tyrosine residues serve as the lism, and tissue homeostasis. -

The Emerging Role of the FGF/FGFR Pathway in Gastrointestinal Stromal Tumor
International Journal of Molecular Sciences Review The Emerging Role of the FGF/FGFR Pathway in Gastrointestinal Stromal Tumor Annalisa Astolfi 1 , Maria Abbondanza Pantaleo 2,*, Valentina Indio 3 , Milena Urbini 4 and Margherita Nannini 5 1 Department of Morphology, Surgery and Experimental Medicine, University of Ferrara, 44121 Ferrara, Italy; annalisa.astolfi@unife.it 2 Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, 40138 Bologna, Italy 3 “Giorgio Prodi” Cancer Research Center, University of Bologna, 40138 Bologna, Italy; [email protected] 4 Biosciences Laboratory, Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST) IRCCS, 47014 Meldola, Italy; [email protected] 5 Medical Oncology Unit, S.Orsola-Malpighi University Hospital, 40138 Bologna, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-051-214-4043; Fax: +39-051-349-655 Received: 10 April 2020; Accepted: 4 May 2020; Published: 7 May 2020 Abstract: Gastrointestinal stromal tumors (GIST) are rare neoplasms of mesenchymal origin arising in the gastrointestinal tract. The vast majority are characterized by mutually exclusive activating mutations in KIT or Platelet-derived growth factor alpha (PDGFRA) receptors, or less frequently by succinate dehydrogenase complex (SDH) or NF1 inactivation, with very rare cases harboring mutant BRAF or RAS alleles. Approximately 5% of GISTs lack any of such mutations and are called quadruple wild-type (WT) GISTs. Recently, deregulated Fibroblast Growth Factor (FGF)/FGF-receptor (FGFR) signaling emerged as a relevant pathway driving oncogenic activity in different molecular subgroups of GISTs. This review summarizes all the current evidences supporting the key role of the FGF/FGFR pathway activation in GISTs, whereby either activating mutations, oncogenic gene fusions, or autocrine/paracrine signaling have been detected in quadruple WT, SDH-deficient, or KIT-mutant GISTs. -

FGF4 Retrogene
PCR Genotyping of a Novel FGF4 Retrogene in Pit Bull Terriers and Mixed Breed Dogs Affected by Disc Disease KA Maciejczyk1, KL Batcher1, DL Bannasch1 1Department of Population Health and Reproduction, School of Veterinary Medicine, University of California-Davis Introduction FGF4 Retrogene Intervertebral Disc Disease (IVDD) is an incredibly devastating and painful disease that affects a wide variety of dog breeds. Chondrodystrophic dog breeds such as Dachshunds and French Bulldogs exhibit the highest frequency of IVDD [1]. Chondrodystrophy is characterized by shortened long bones and prematurely calcified intervertebral discs [2]. IVDD is the result of herniation of this intervertebral disc into the spinal canal often causing pain and paralysis. A recently discovered fibroblast growth factor 4 (FGF4) retrogene on chromosome (Chr) 12 is responsible for canine chondrodystrophy and a leading contributor to a majority of IVDD cases [3]. However, not all dogs affected with IVDD have the Chr 12 retrogene [Brown et al. and K. Batcher and D. Bannasch unpublished results]. Recently the laboratory identified a second FGF4 The FGF4 retrogene on Chr 13 came from the endogenous FGF4 gene on chromosome 18. FGF4 is composed of retrogene on Chr 13 in two IVDD affected three exons and two introns. When FGF4 is transcribed the introns are removed and a poly-A tail is added. This Pit Bull Terriers that did not have the Chr 12 entire transcript is approximately 3.2kb long. Retrogene formation is a rare event that can occur when RNA is retrogene. The purpose of this study was to reverse transcribed into DNA and integrated back elsewhere in the genome [4]. -

Epha2 and EGFR: Friends in Life, Partners in Crime
cancers Review EphA2 and EGFR: Friends in Life, Partners in Crime. Can EphA2 Be a Predictive Biomarker of Response to Anti-EGFR Agents? Mario Cioce 1,* and Vito Michele Fazio 1,2,3,* 1 Laboratory of Molecular Medicine and Biotechnology, Department of Medicine, University Campus Bio-Medico of Rome, 00128 Rome, Italy 2 Laboratory of Oncology, Fondazione IRCCS Casa Sollievo della Sofferenza, 71013 San Giovanni Rotondo, Italy 3 Institute of Translational Pharmacology, National Research Council of Italy (CNR), 00133 Rome, Italy * Correspondence: [email protected] (M.C.); [email protected] (V.M.F.) Simple Summary: The Ephrin receptors and their ligands play important roles in organ formation and tissue repair, by orchestrating complex programs of cell adhesion and repulsion, however, this same system plays a role in cancer development In fact, EphA2 levels are higher in tumors vs normal tissue and further increased upon treatment, in vivo and in vitro. Changes in the molecular status of EphA2, of its subcellular localization, the absence of ligand and signals derived from the tumor context unleash the oncogenic role of EphA2 and its broad ability to promote resistance to radiotherapy, chemotherapy and targeted agents, including inhibitors of Epidermal-Growth-Factor- Receptor (EGFR). High levels of EphA2 may reduce response to cetuximab even in RAS wt CRC patients. In this work, we aim to review the current knowledge of the EphA2 function which is crucial for achieving a more effective therapeutic management of tumors resistant to EGFR inhibitors and to many other agents. Abstract: The Eph receptors represent the largest group among Receptor Tyrosine kinase (RTK) Citation: Cioce, M.; Fazio, V.M. -

FGFR4 Inhibitor BLU9931 Attenuates Pancreatic Cancer Cell Proliferation and Invasion While Inducing Senescence
cancers Article FGFR4 Inhibitor BLU9931 Attenuates Pancreatic Cancer Cell Proliferation and Invasion While Inducing Senescence: Evidence for Senolytic Therapy Potential in Pancreatic Cancer Norihiko Sasaki 1, Fujiya Gomi 2, Hisashi Yoshimura 3 , Masami Yamamoto 3, Yoko Matsuda 4 , Masaki Michishita 5, Hitoshi Hatakeyama 6, Yoichi Kawano 7 , Masashi Toyoda 1 , Murray Korc 8 and Toshiyuki Ishiwata 2,* 1 Research team for Geriatric Medicine (Vascular Medicine), Tokyo Metropolitan Institute of Gerontology, Sakae-cho 35-2, Itabashi-ku, Tokyo 173-0015, Japan; [email protected] (N.S.); [email protected] (M.T.) 2 Division of Aging and Carcinogenesis, Research Team for Geriatric Pathology, Tokyo Metropolitan Institute of Gerontology, Tokyo 173-0015, Japan; [email protected] 3 Division of Physiological Pathology, Department of Applied Science, School of Veterinary Nursing and Technology, Nippon Veterinary and Life Science University, Tokyo 180-8602, Japan; [email protected] (H.Y.); [email protected] (M.Y.) 4 Oncology Pathology, Department of Pathology and Host-Defense, Kagawa University, Kagawa 761-0793, Japan; [email protected] 5 Department of Veterinary Pathology, School of Veterinary Medicine, Nippon Veterinary and Life Science University, Tokyo 180-8602, Japan; [email protected] 6 Department of Comprehensive Education in Veterinary Medicine, Nippon Veterinary and Life Science University, Tokyo 180-8602, Japan; [email protected] 7 Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Nippon Medical School, Tokyo 113-8603, Japan; [email protected] 8 Department of Developmental and Cell Biology, School of Biological Sciences, University of California, Irvine, CA 92697, USA; [email protected] * Correspondence: [email protected]; Tel.: +81-3-3964-1141 (ext.