Genome Mining, Heterologous Expression, Antibacterial and Antioxidant Activities of Lipoamides and Amicoumacins from Compost-Associated
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Monitoring Suaeda Salsa Spectral Response to Salt Conditions in Coastal Wetlands: a Case Study in Dafeng Elk National Nature Reserve, China
remote sensing Article Monitoring Suaeda salsa Spectral Response to Salt Conditions in Coastal Wetlands: A Case Study in Dafeng Elk National Nature Reserve, China Xia Lu 1,2, Sen Zhang 1,2, Yanqin Tian 1,2, Yurong Li 1,2, Rui Wen 3, JinYau Tsou 4,5 and Yuanzhi Zhang 5,6,* 1 Jiangsu Key laboratory of Marine Bioresources and Environment/Jiangsu Key laboratory of Marine Biotechnology/Co-Innovation Center of Jiangsu Marine Bio-Industry Technology, Jiangsu Ocean University, Lianyungang 222005, China; [email protected] (X.L.); [email protected] (S.Z.); [email protected] (Y.T.); [email protected] (Y.L.) 2 School of Geomatics and Marine Information, Jiangsu Ocean University, Lianyungang 222005, China 3 Third Institute of Oceanography, Ministry of Natural Resources, Xiamen 361005, China; [email protected] 4 Department of Architecture and Civil Engineering, City University of Hong Kong, Hong Kong, China; [email protected] 5 Faculty of Social Science and Asia-Pacific Studies Institute, Chinese University of Hong Kong, Hong Kong, China 6 School of Marine Science, Nanjing University of Information Science and Technology, Nanjing 210044, China * Correspondence: [email protected]; Tel.: +86-188-8885-3470 Received: 17 July 2020; Accepted: 17 August 2020; Published: 20 August 2020 Abstract: This paper reports on monitored Suaeda salsa spectral response to salt conditions in coastal wetlands, using spectral measurements and remotely sensed algorithms. Suaeda salsa seedlings were collected from the Dafeng Elk National Nature Reserve (DENNR) in Jiangsu Province, China. We treated 21 Suaeda salsa seedlings planted in pots with 7 different salt concentrations (n = 3 for each concentration) to assess their response to varying salt conditions. -

Using Macroalgae As Biofuel: Current Opportunities and Challenges
Botanica Marina 2020; aop Review Guang Gao*, James Grant Burgess, Min Wu, Shujun Wang and Kunshan Gao* Using macroalgae as biofuel: current opportunities and challenges https://doi.org/10.1515/bot-2019-0065 Received 9 September, 2019; accepted 18 February, 2020 Introduction Abstract: The rising global demand for energy and the Today, approximately 85% of the total energy consumed decreasing stocks of fossil fuels, combined with envi- worldwide is provided by fossil fuels (Dudley 2018). Stocks ronmental problems associated with greenhouse gas of fossil fuels are declining, and there is growing concern emissions, are driving research and development for regarding the serious environmental problems associated alternative and renewable sources of energy. Algae have with their consumption (Ravi et al. 2018, Saratale et al. been gaining increasing attention as a potential source 2018). Consequently, it is of general importance to search of bio-renewable energy because they grow rapidly, and for renewable and cost-effective energy sources with low or farming them does not, generally, compete for agricul- zero greenhouse gas emissions (Ravi et al. 2018). Biofuels, tural land use. Previous studies of algal biofuels have mainly extracted from plants, can serve as an attractive focused on microalgae because of their fast growth rate source of energy to meet some of the present and future and high lipid content. Here we analyze the multiple mer- fuel demand (Robertson et al. 2017). Currently, bio- ethanol its of biofuel production using macroalgae, with particu- and biodiesel, which are produced primarily from food lar reference to their chemical composition, biomass and crops such as grain, sugar cane and vegetable oils, are the biofuel productivity, and cost-effectiveness. -

A Complete Collection of Chinese Institutes and Universities For
Study in China——All China Universities All China Universities 2019.12 Please download WeChat app and follow our official account (scan QR code below or add WeChat ID: A15810086985), to start your application journey. Study in China——All China Universities Anhui 安徽 【www.studyinanhui.com】 1. Anhui University 安徽大学 http://ahu.admissions.cn 2. University of Science and Technology of China 中国科学技术大学 http://ustc.admissions.cn 3. Hefei University of Technology 合肥工业大学 http://hfut.admissions.cn 4. Anhui University of Technology 安徽工业大学 http://ahut.admissions.cn 5. Anhui University of Science and Technology 安徽理工大学 http://aust.admissions.cn 6. Anhui Engineering University 安徽工程大学 http://ahpu.admissions.cn 7. Anhui Agricultural University 安徽农业大学 http://ahau.admissions.cn 8. Anhui Medical University 安徽医科大学 http://ahmu.admissions.cn 9. Bengbu Medical College 蚌埠医学院 http://bbmc.admissions.cn 10. Wannan Medical College 皖南医学院 http://wnmc.admissions.cn 11. Anhui University of Chinese Medicine 安徽中医药大学 http://ahtcm.admissions.cn 12. Anhui Normal University 安徽师范大学 http://ahnu.admissions.cn 13. Fuyang Normal University 阜阳师范大学 http://fynu.admissions.cn 14. Anqing Teachers College 安庆师范大学 http://aqtc.admissions.cn 15. Huaibei Normal University 淮北师范大学 http://chnu.admissions.cn Please download WeChat app and follow our official account (scan QR code below or add WeChat ID: A15810086985), to start your application journey. Study in China——All China Universities 16. Huangshan University 黄山学院 http://hsu.admissions.cn 17. Western Anhui University 皖西学院 http://wxc.admissions.cn 18. Chuzhou University 滁州学院 http://chzu.admissions.cn 19. Anhui University of Finance & Economics 安徽财经大学 http://aufe.admissions.cn 20. Suzhou University 宿州学院 http://ahszu.admissions.cn 21. -

Research on the Majors Setting in Colleges and Universities and Its Dynamic Adjustment Mechanism Based on Regional Economic Deve
2017 2nd International Conference on Modern Economic Development and Environment Protection (ICMED 2017) ISBN: 978-1-60595-518-6 Research on the Majors Setting in Colleges and Universities and Its Dynamic Adjustment Mechanism Based on Regional Economic Development—A Case Study of Jiangsu Province Ming CHEN1,a, Hua-liang LU1,b,* and Cheng ZHANG1,c 1Teaching Affairs Office, Nanjing University of Finance and Economics, Nanjing, China [email protected], [email protected], [email protected] *Corresponding author Keywords: Regional Economic Development, Majors Setting in Colleges and Universities, Dynamic Adjustment Mechanism. Abstract. Whether majors setting in colleges and universities can meet the human resource demands of regional economic development is related not only to the transformation of current regional economy and the adjustment of industrial structure, but also to the functional attributes of colleges and universities and their compliance with the requirements of “Double First-rate” construction in the reform of higher education. However, there are three major problems in the current majors setting of colleges and universities: firstly, there is short of overall planning and the homogenization is serious; another problem is that majors setting are arbitrary and blind; thirdly, professional division is too far elaborate, resulting in the narrow employment opportunities. Based on the problems of majors setting in colleges and universities and their properties, we put forward that it is necessary to establish the information sharing of majors in regional colleges and universities to solve the problem of information asymmetry, which is conducive to adjusting majors setting in time, thus forming a dynamic adjustment mechanism. 1. -

University of Leeds Chinese Accepted Institution List 2021
University of Leeds Chinese accepted Institution List 2021 This list applies to courses in: All Engineering and Computing courses School of Mathematics School of Education School of Politics and International Studies School of Sociology and Social Policy GPA Requirements 2:1 = 75-85% 2:2 = 70-80% Please visit https://courses.leeds.ac.uk to find out which courses require a 2:1 and a 2:2. Please note: This document is to be used as a guide only. Final decisions will be made by the University of Leeds admissions teams. -
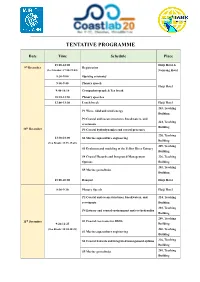
Tentative Programme
TENTATIVE PROGRAMME Date Time Schedule Place 15:00-22:00 Haiji Hotel & 9th December Registration (Ice breaker: 17:00-19:00) Nanyang Hotel 8:30-9:00 Opening ceremony 9:00-9:40 Plenary speech Haiji Hotel 9:40-10:10 Group photograph & Tea break 10:10-11:50 Plenary speeches 12:00-13:30 Lunch break Haiji Hotel 203, Teaching P1 Wave, tidal and wind energy Building P2 Coastal and ocean structures, breakwaters, and 224, Teaching revetments th Building 10 December P3 Coastal hydrodynamics and coastal processes 220, Teaching 13:30-18:00 S2 Marine aquaculture engineering Building (Tea Break: 15:25-15:45) 209, Teaching S3 Evolution and modeling of the Yellow River Estuary Building S4 Coastal Hazards and Integrated Management 226, Teaching Options Building 201, Teaching S5 Marine geotechnics Building 18:00-20:00 Banquet Haiji Hotel 8:30-9:10 Plenary Speech Haiji Hotel P2 Coastal and ocean structures, breakwaters, and 224, Teaching revetments Building 203, Teaching P4 Estuary and coastal environment and eco-hydraulics Building 209, Teaching 11th December S1 Coastal reservoirs for SDG6 9:20-12:25 Building (Tea Break: 10:30-10:45) 220, Teaching S2 Marine aquaculture engineering Building S4 Coastal hazards and integrated management options 226, Teaching Building S5 Marine geotechnics 201, Teaching Building 12:00-13:30 Lunch break Haiji Hotel 13:30-17:30 Technical tour 17:30-19:30 Dinner Haiji Hotel 224, Teaching P3 Coastal hydrodynamics and coastal processes Building 201, Teaching P5 Laboratory techniques and measurement systems Building P6 Field measurement, -

Delegates Organization Background Beijing Hydrosurvey Science
The 4th China Ocean Technology Conference M ay 16 - 18, 2019, Zhoushan Campus, Zhejiang University D elegates Organization Background Beijing Hydrosurvey Science & Technology Co., Ltd Beijing Institute of Exploration Engineering, MNR Beijing Normal University BGP Inc., China National Petroleum Corporation CETC Chongqing Acoustic - Optic - Electronic Co., Ltd. Chang An University Changshu Ruite Electric Co., Ltd. China Geological Survey China Huarong Ocean Fisheries Development Co., Ltd. China Jiliang University China Oceanic Information Network China University of Geosciences, Beijing China University of Geosciences, Wuhan China University of Petroleum China University of Petroleum (Eastern China) China Academy of Electronics and Information Technology Chinese Institute of Electronics CIMC Ocean Engineering Institute, Zhejiang University CNOOC Research Institute College of Electrical Engineering, Zhejiang University College of Geophysics, China University of Petroleum College of Information Science & Electronic Engineering, Zhejiang University College of Ocean and Earth Sciences, Xiamen University College of Oceanography, Hohai University Dalian University of Technology Department of Physics, Ocean University of China East China Normal University Fudan University Gaungxi University Geological Exploration Technology Institute of Jiangsu Province Guangdong Sea Star Ocean Sci. and Tech. Co., Ltd. Guangzhou Institute of Energy Conversion, Chinese Academy of Sciences Guangzhou Marine Geological Survey Guodian -

Study on Desertification Reversal Factors in Maowusu Sandy Land in China
E3S Web of Conferences 199, 00015 (2020) https://doi.org/10.1051/e3sconf/202019900015 ICWREE2020 Study on desertification reversal factors in Maowusu sandy land in China Zhang Ying1, Lijun Wang2, AiJun Yi3, Fei Li4, and Ernst-August Nuppenau5 1School of Economics & Management, Beijing Forestry University, Beijing 100083,China 2School of Economics and Management, Nanjing University of Aeronautics and Astronautics,Nanjing 211106, China 3School of Business, Jiangsu Ocean University, Lianyungang 222005, China 4School of Landscape Architecture, Beijing Forestry University, Beijing 100083,China 5Institute of Agricultural Politics and Monetary Research, Giessen University, D-35390 Giessen, Germany Abstract: Based on the data of China's economic and social big data platform from 2000 to 2019, this paper studies and analyzes the development status and influencing factors of desertification in Maowusu sandy land in China. Based on the exploratory analysis (EFA) and the dummy variable regression model (DVRM), the research result shows that the annual precipitation is the main climate factor affecting the vegetation coverage in this area. And for every 100 mm increase in annual precipitation, vegetation coverage will increase by 10%. In addition, the annual average temperature also has a significant impact on the vegetation coverage. For every 1 ℃ increase in the annual average temperature, the vegetation coverage will increase by 2.5%. Analysis of policy factors shows that the policy effects of the 2005 National Desert Control Plan (2005-2010) and the 2011 National Desert Control Plan (2011-2020) etc. can increase vegetation coverage by 3.4% and 4.7%respectively compared with the base period level in 2000. The study reveals the important role of climate and policy factors in the reversion of desertification in Maowusu sandy land. -

Friday, November 1 , 2019 Saturday, November 2 , 2019 08:30-12:00
FRIDAY, NOVEMBER 1st, 2019 09:00-20:30 Arrival & Registration 18:00-20:00 Dinner SATURDAY, NOVEMBER 2nd, 2019 08:30-12:00 Building No. 3, Meeting Room No. 1 Opening Ceremony 08:30-08:55 Chaired by Guochen Zhang (Vice President of Dalian Ocean University) Welcoming Address by Jie Yao (Chairman of Dalian Ocean University 08:30-08:35 Council) Address by Daigang Yang (Chairman of Dalian Association for Science and 08:35-08:40 Technology Council) Address by L. Courtney Smith (President of International Society of 08:40-08:45 Developmental and Comparative Immunology) Address by Linsheng Song (President of Asian Society of Developmental and 08:45-08:50 Comparative Immunology) Conferring the Certificate of Honorary President and Vice President of 08:50-08:55 ASDCI 08:55-09:10 Group Photo Plenary Lecture 09:10-12:00 Chaired by Linsheng Song and Miki Nakao L. Courtney Smith (George Washington University, USA) 09:10-09:50 Sequence diversity in the genes and proteins of the SpTransformer system in sea urchins Kenneth Söderhäll 09:50-10:30 (Uppsala University, Sweden) Blood cell synthesis in a crustacean 10:30-10:40 Break Yaofeng Zhao (China Agricultural University, China) 10:40-11:20 Immunoglobulins in tetrapod other than human and mouse: surprises and applications Maria Forlenza 11:20-12:00 (Wageningen University, Netherlands) Trypanosome infections in Zebrafish: seeing is believing 12:00 Lunch SATURDAY, NOVEMBER 2nd, 2019 14:00-17:35 Building No. 5, Building No. 5, Building No. 1, Room 402 Multi-function Hall Meeting Room No. 5 Session 1 Session 2 Session 3 Evolution and origins of Innate Immunity Ecoimmunology and immune system immunometabolism Chaired by L. -

Program and Abstract
Online International Symposium of FSMILE 2020 Nov 24th-25th, 2020 Venue: Meeting room of each organization connected by Zoom Online Zoom ID: 659 283 7473 (Time zone: GMT+9:00, Tokyo time) Nov 24h Morning Session:8:30-12:00 Chair: Dr. Chunhong YUAN (Iwate University), Dr. Hong-shun YANG (National University of Singapore) 8 : 30 - 8 : 45 online registration Opening remarks ( Satoshi OGAWA, President, Iwate University) 8 : 45 - 9 : 00 (Noriyuki TANAKA, Vice-Director, NPO FSMILE) Plant Polyphenols as Natural Additives to Inhibit Oxidation and Mitigate Toxicants 9 : 00 - 9 : 35 KS 1 Youling L. XIONG Professor University of Kentucky in Muscle Foods DEVELOPMENT OF PROCESSING TECHNOLOGY OF SEAFOOD 9 : 35 - 10 : 10 KS 2 Emiko OKAZAKI Professor Tokyo University of Marine Science and Technology IN JAPAN 10 : 10 - 10 : 35 IS 1 Hong-shun YANG Associate Professor National University of Singapore Application of metabolomics in seafood quality and microbial safety 10 : 35 - 11 : 00 IS 2 Yuya KUMAGAI Assistant Professor Hokkaido University Healthy functional materials in red algae Efficient extraction and monthly variation of mycosporine-like amino acids from 11 : 00 - 11 : 15 O 1 Yuki NISHIDA M1 Hokkaido University red alga dulse in Japan Functional characteristics improvement of HPMC modified PVA film incorporating 11 : 15 - 11 : 30 O 2 Jiayin HUANG M3 Zhejiang University roselle anthocyanins for shrimp freshness monitoring Evaluation of beef (Wenling-Gaofeng cattle) tenderness during Sous vide cooking: 11 : 30 - 11 : 45 O 3 Meiyu CHEN M3 Zhejiang University a water phase transition and myofibrillar protein structural perspective Effects of sterilization pretreatment combined with natural chemicals and/or ultra- 11 : 45 - 12 : 00 O 4 Sijia Peng M3 Nanjing Normal University high pressure on the quality and safety of pickled raw mud snails (Bullacta exarata) after storage at −20°C Food and Break Nov 24h afternoon Session:13:00-18:00 Chair: Prof. -

2021 MCM Contest
2021 Mathematical Contest in Modeling® Press Release—April 23, 2021 COMAP is pleased to announce the results of the 37th annual advantages and disadvantages for fungi species and combinations of Mathematical Contest in Modeling (MCM). This year, 10,053 teams species like to persist is various environments. representing institutions from fifteen countries/regions participated in the contest. Seventeen teams from the following institutions were The B problem used the scenario of the 2019-2020 fire season in designated as OUTSTANDING WINNERS: Australia, which saw devastating wildfires in every state, to consider the use of drones in firefighting. Teams learned of the capabilities of Shanghai Jiao Tong University, China (3) two types of drones, surveillance and situational awareness drones and (2100454 SIAM Award & COMAP Scholarship Award) hovering drones that can carry repeaters (to extend radio range), and Beijing Institute of Technology, China (Ben Fusaro Award) then created a model to determine the optimal numbers and mix of Nanjing University of Posts & Telecommunications, China these two types of drones. Teams addressed adaptation of their model (Frank Giordano Award & SIAM Award) to the changing likelihood of extreme fire events over the next decade, Jiangnan University, China as well as to equipment cost increases. Teams also developed a model Xi'an Jiaotong University, China (AMS Award) to optimize locations of hovering drones for fires of differing sizes on Xidian University, Shannxi, China differing terrain. Beijing Jiaotong University, China (ASA Award) University of Colorado Boulder, CO, USA The C problem investigated the discovery and sightings of Vespa (MAA Award, SIAM Award & COMAP Scholarship Award) mandarina (also known as the Asian giant hornet) in the State of University of Oxford, United Kingdom Washington. -

Rapid Determination of Iodine Content in Drinking Water by Isopropyl Sensitization and Inductively Coupled Plasma Mass Spectrometry (ICP-MS)
E3S Web of Conferences , 03004 (2021) https://doi.org/10.1051/e3sconf/202124503004 AEECS 2021 Rapid determination of iodine content in drinking water by isopropyl sensitization and inductively coupled plasma mass spectrometry (ICP-MS) Yanxia Zeng1,2, Xiashi Zhu1* 1Department of Chemical Engineering, Yangzhou University,Yangzhou City, Jiangsu Province漓225009漓China 2Institute of Marine Resource Development漓Jiangsu Ocean University, Lianyungang City, Jiangsu Province漓222005漓China) Abstract. A subject for the rapid determination of iodine content in drinking water by isopropyl sensitization and ICP-MS was established. The samples were dissolved in 1% ammonia solution,127I was selected as the mass number to be measured and 159Tb as an internal standard. At the same time, 4% (V/V) isopropanol was used to enhance aerosol gasification efficiency, 2% NH3·H2O was used to effectively eliminate memory effects of iodine by cleaning sample for 180 seconds. The results show that, the detection limits of iodine in this method are 0.007 mg·kg-1 and it is superior to other literature.The standard curve has a good linear relationship and the recovery rates of testing rusults range are from 96.00% to 112.3%. The content of iodine in standard substance water samples was determined by ICP-MS method. The iodine content in water samples from seven drinking water sources in Lianyungang area was analyzed between 14~21 μg·L-1. It was in line with the allowable value of 10~300 μg·L-1 of the sanitary standard for drinking water, suggesting that the iodine content of drinking water resources in Lianyungang area was in a safe range.