Introduction to Data Mining with R
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Columbus Regional Airport Authority
COLUMBUS REGIONAL AIRPORT AUTHORITY - PORT COLUMBUS INTERNATIONAL AIRPORT TRAFFIC REPORT June 2014 7/22/2014 Airline Enplaned Passengers Deplaned Passengers Enplaned Air Mail Deplaned Air Mail Enplaned Air Freight Deplaned Air Freight Landings Landed Weight Air Canada Express - Regional 2,377 2,278 - - - - 81 2,745,900 Air Canada Express Totals 2,377 2,278 - - - - 81 2,745,900 AirTran 5,506 4,759 - - - - 59 6,136,000 AirTran Totals 5,506 4,759 - - - - 59 6,136,000 American 21,754 22,200 - - - 306 174 22,210,000 Envoy Air** 22,559 22,530 - - 2 ,027 2 ,873 527 27,043,010 American Totals 44,313 44,730 - - 2,027 3,179 701 49,253,010 Delta 38,216 36,970 29,594 34,196 25,984 36,845 278 38,899,500 Delta Connection - ExpressJet 2,888 2,292 - - - - 55 3,709,300 Delta Connection - Chautauqua 15,614 14,959 - - 640 - 374 15,913,326 Delta Connection - Endeavor 4 ,777 4,943 - - - - 96 5,776,500 Delta Connection - GoJet 874 748 - - 33 - 21 1,407,000 Delta Connection - Shuttle America 6,440 7,877 - - 367 - 143 10,536,277 Delta Connection - SkyWest 198 142 - - - - 4 188,000 Delta Totals 69,007 67,931 29,594 34,196 27,024 36,845 971 76,429,903 Southwest 97,554 96,784 218,777 315,938 830 103,146,000 Southwest Totals 97,554 96,784 - - 218,777 315,938 830 103,146,000 United 3 ,411 3,370 13,718 6 ,423 1 ,294 8 ,738 30 3,990,274 United Express - ExpressJet 13,185 13,319 - - - - 303 13,256,765 United Express - Mesa 27 32 - - - - 1 67,000 United Express - Republic 4,790 5,133 - - - - 88 5,456,000 United Express - Shuttle America 9,825 9,076 - - - - 151 10,919,112 -
Automated Flight Statistics Report For
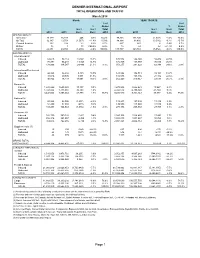
DENVER INTERNATIONAL AIRPORT TOTAL OPERATIONS AND TRAFFIC March 2014 March YEAR TO DATE % of % of % Grand % Grand Incr./ Incr./ Total Incr./ Incr./ Total 2014 2013 Decr. Decr. 2014 2014 2013 Decr. Decr. 2014 OPERATIONS (1) Air Carrier 36,129 35,883 246 0.7% 74.2% 99,808 101,345 (1,537) -1.5% 73.5% Air Taxi 12,187 13,754 (1,567) -11.4% 25.0% 34,884 38,400 (3,516) -9.2% 25.7% General Aviation 340 318 22 6.9% 0.7% 997 993 4 0.4% 0.7% Military 15 1 14 1400.0% 0.0% 18 23 (5) -21.7% 0.0% TOTAL 48,671 49,956 (1,285) -2.6% 100.0% 135,707 140,761 (5,054) -3.6% 100.0% PASSENGERS (2) International (3) Inbound 68,615 58,114 10,501 18.1% 176,572 144,140 32,432 22.5% Outbound 70,381 56,433 13,948 24.7% 174,705 137,789 36,916 26.8% TOTAL 138,996 114,547 24,449 21.3% 3.1% 351,277 281,929 69,348 24.6% 2.8% International/Pre-cleared Inbound 42,848 36,668 6,180 16.9% 121,892 102,711 19,181 18.7% Outbound 48,016 39,505 8,511 21.5% 132,548 108,136 24,412 22.6% TOTAL 90,864 76,173 14,691 19.3% 2.0% 254,440 210,847 43,593 20.7% 2.1% Majors (4) Inbound 1,698,200 1,685,003 13,197 0.8% 4,675,948 4,662,021 13,927 0.3% Outbound 1,743,844 1,713,061 30,783 1.8% 4,724,572 4,700,122 24,450 0.5% TOTAL 3,442,044 3,398,064 43,980 1.3% 75.7% 9,400,520 9,362,143 38,377 0.4% 75.9% National (5) Inbound 50,888 52,095 (1,207) -2.3% 139,237 127,899 11,338 8.9% Outbound 52,409 52,888 (479) -0.9% 139,959 127,940 12,019 9.4% TOTAL 103,297 104,983 (1,686) -1.6% 2.3% 279,196 255,839 23,357 9.1% 2.3% Regionals (6) Inbound 382,759 380,328 2,431 0.6% 1,046,306 1,028,865 17,441 1.7% Outbound -

Columbus Regional Airport Authority
COLUMBUS REGIONAL AIRPORT AUTHORITY - PORT COLUMBUS INTERNATIONAL AIRPORT TRAFFIC REPORT October, 2009 11/24/2009 Airline Enplaned Passengers Deplaned Passengers Enplaned Air Mail Deplaned Air Mail Enplaned Air Freight Deplaned Air Freight Landings Landed Weight Air Canada Jazz - Regional 1,385 1,432 0 0 0 0 75 2,548,600 Air Canada Jazz Totals 1,385 1,432 0 0 0 0 75 2,548,600 AirTran 16,896 16,563 0 0 0 0 186 20,832,000 AirTran Totals 16,896 16,563 0 0 0 0 186 20,832,000 American 13,482 13,047 10,256 13,744 0 75 120 14,950,000 American Connection - Chautauqua 0 0 0 0 0 0 0 0 American Eagle 22,258 22,818 0 0 2,497 3,373 550 24,434,872 American Totals 35,740 35,865 10,256 13,744 2,497 3,448 670 39,384,872 Continental 5,584 5,527 24,724 17,058 6,085 12,750 57 6,292,000 Continental Express - Chautauqua 4,469 4,675 0 0 477 0 110 4,679,500 Continental Express - Colgan 2,684 3,157 0 0 0 0 69 4,278,000 Continental Express - CommutAir 1,689 1,630 0 0 0 0 64 2,208,000 Continental Express - ExpressJet 3,821 3,334 0 0 459 1,550 100 4,122,600 Continental Totals 18,247 18,323 24,724 17,058 7,021 14,300 400 21,580,100 Delta 14,640 13,970 0 0 9,692 38,742 119 17,896,000 Delta Connection - Atlantic SE 2,088 2,557 0 1 369 2 37 2,685,800 Delta Connection - Chautauqua 13,857 13,820 0 0 0 0 359 15,275,091 Delta Connection - Comair 1,890 1,802 0 0 0 0 52 2,444,000 Delta Connection - Mesa/Freedom 0 0 0 0 0 0 0 0 Delta Connection - Pinnacle 0 0 0 0 0 0 0 0 Delta Connection - Shuttle America 4,267 4,013 0 0 0 0 73 5,471,861 Delta Connection - SkyWest 0 0 0 0 -

Bankruptcy Tilts Playing Field Frank Boroch, CFA 212 272-6335 [email protected]
Equity Research Airlines / Rated: Market Underweight September 15, 2005 Research Analyst(s): David Strine 212 272-7869 [email protected] Bankruptcy tilts playing field Frank Boroch, CFA 212 272-6335 [email protected] Key Points *** TWIN BANKRUPTCY FILINGS TILT PLAYING FIELD. NWAC and DAL filed for Chapter 11 protection yesterday, becoming the 20 and 21st airlines to do so since 2000. Now with 47% of industry capacity in bankruptcy, the playing field looks set to become even more lopsided pressuring non-bankrupt legacies to lower costs further and low cost carriers to reassess their shrinking CASM advantage. *** CAPACITY PULLBACK. Over the past 20 years, bankrupt carriers decreased capacity by 5-10% on avg in the year following their filing. If we assume DAL and NWAC shrink by 7.5% (the midpoint) in '06, our domestic industry ASM forecast goes from +2% y/y to flat, which could potentially be favorable for airline pricing (yields). *** NWAC AND DAL INTIMATE CAPACITY RESTRAINT. After their filing yesterday, NWAC's CEO indicated 4Q:05 capacity could decline 5-6% y/y, while Delta announced plans to accelerate its fleet simplification plan, removing four aircraft types by the end of 2006. *** BIGGEST BENEFICIARIES LIKELY TO BE LOW COST CARRIERS. NWAC and DAL account for roughly 26% of domestic capacity, which, if trimmed by 7.5% equates to a 2% pt reduction in industry capacity. We believe LCC-heavy routes are likely to see a disproportionate benefit from potential reductions at DAL and NWAC, with AAI, AWA, and JBLU in particular having an easier path for growth. -

U.S. Department of Transportation Federal
U.S. DEPARTMENT OF ORDER TRANSPORTATION JO 7340.2E FEDERAL AVIATION Effective Date: ADMINISTRATION July 24, 2014 Air Traffic Organization Policy Subject: Contractions Includes Change 1 dated 11/13/14 https://www.faa.gov/air_traffic/publications/atpubs/CNT/3-3.HTM A 3- Company Country Telephony Ltr AAA AVICON AVIATION CONSULTANTS & AGENTS PAKISTAN AAB ABELAG AVIATION BELGIUM ABG AAC ARMY AIR CORPS UNITED KINGDOM ARMYAIR AAD MANN AIR LTD (T/A AMBASSADOR) UNITED KINGDOM AMBASSADOR AAE EXPRESS AIR, INC. (PHOENIX, AZ) UNITED STATES ARIZONA AAF AIGLE AZUR FRANCE AIGLE AZUR AAG ATLANTIC FLIGHT TRAINING LTD. UNITED KINGDOM ATLANTIC AAH AEKO KULA, INC D/B/A ALOHA AIR CARGO (HONOLULU, UNITED STATES ALOHA HI) AAI AIR AURORA, INC. (SUGAR GROVE, IL) UNITED STATES BOREALIS AAJ ALFA AIRLINES CO., LTD SUDAN ALFA SUDAN AAK ALASKA ISLAND AIR, INC. (ANCHORAGE, AK) UNITED STATES ALASKA ISLAND AAL AMERICAN AIRLINES INC. UNITED STATES AMERICAN AAM AIM AIR REPUBLIC OF MOLDOVA AIM AIR AAN AMSTERDAM AIRLINES B.V. NETHERLANDS AMSTEL AAO ADMINISTRACION AERONAUTICA INTERNACIONAL, S.A. MEXICO AEROINTER DE C.V. AAP ARABASCO AIR SERVICES SAUDI ARABIA ARABASCO AAQ ASIA ATLANTIC AIRLINES CO., LTD THAILAND ASIA ATLANTIC AAR ASIANA AIRLINES REPUBLIC OF KOREA ASIANA AAS ASKARI AVIATION (PVT) LTD PAKISTAN AL-AAS AAT AIR CENTRAL ASIA KYRGYZSTAN AAU AEROPA S.R.L. ITALY AAV ASTRO AIR INTERNATIONAL, INC. PHILIPPINES ASTRO-PHIL AAW AFRICAN AIRLINES CORPORATION LIBYA AFRIQIYAH AAX ADVANCE AVIATION CO., LTD THAILAND ADVANCE AVIATION AAY ALLEGIANT AIR, INC. (FRESNO, CA) UNITED STATES ALLEGIANT AAZ AEOLUS AIR LIMITED GAMBIA AEOLUS ABA AERO-BETA GMBH & CO., STUTTGART GERMANY AEROBETA ABB AFRICAN BUSINESS AND TRANSPORTATIONS DEMOCRATIC REPUBLIC OF AFRICAN BUSINESS THE CONGO ABC ABC WORLD AIRWAYS GUIDE ABD AIR ATLANTA ICELANDIC ICELAND ATLANTA ABE ABAN AIR IRAN (ISLAMIC REPUBLIC ABAN OF) ABF SCANWINGS OY, FINLAND FINLAND SKYWINGS ABG ABAKAN-AVIA RUSSIAN FEDERATION ABAKAN-AVIA ABH HOKURIKU-KOUKUU CO., LTD JAPAN ABI ALBA-AIR AVIACION, S.L. -

Pilots Jump to Each Section Below Contents by Clicking on the Title Or Photo
November 2018 Aero Crew News Your Source for Pilot Hiring and More... ExpressJet is taking off with a new Pilot Contract Top-Tier Compensation and Work Rules $40/hour first-year pay $10,000 annual override for First Officers, $8,000 for Captains New-hire bonus 100% cancellation and deadhead pay $1.95/hour per-diem Generous 401(k) match Friendly commuter and reserve programs ARE YOU READY FOR EXPRESSJET? FLEET DOMICILES UNITED CPP 126 - Embraer ERJ145 Chicago • Cleveland Spend your ExpressJet career 20 - Bombardier CRJ200 Houston • Knoxville knowing United is in Newark your future with the United Pilot Career Path Program Apply today at expressjet.com/apply. Questions? [email protected] expressjet.com /ExpressJetPilotRecruiting @expressjetpilots Jump to each section Below contents by clicking on the title or photo. November 2018 20 36 24 50 32 Also Featuring: Letter from the Publisher 8 Aviator Bulletins 10 Self Defense for Flight Crews 16 Trans States Airlines 42 4 | Aero Crew News BACK TO CONTENTS the grid New Airline Updated Flight Attendant Legacy Regional Alaska Airlines Air Wisconsin The Mainline Grid 56 American Airlines Cape Air Delta Air Lines Compass Airlines Legacy, Major, Cargo & International Airlines Hawaiian Airlines Corvus Airways United Airlines CommutAir General Information Endeavor Air Work Rules Envoy Additional Compensation Details Major ExpressJet Airlines Allegiant Air GoJet Airlines Airline Base Map Frontier Airlines Horizon Air JetBlue Airways Island Air Southwest Airlines Mesa Airlines Spirit Airlines -

Customers First Plan, Highlighting Definitions of Terms
RepLayout for final pdf 8/28/2001 9:24 AM Page 1 2001 Annual Report [c u s t o m e r s] AIR TRANSPORT ASSOCIATION RepLayout for final pdf 8/28/2001 9:24 AM Page 2 Officers Carol B. Hallett President and CEO John M. Meenan Senior Vice President, Industry Policy Edward A. Merlis Senior Vice President, Legislative and International Affairs John R. Ryan Acting Senior Vice President, Aviation Safety and Operations Vice President, Air Traffic Management Robert P. Warren mi Thes Air Transports i Associationo n of America, Inc. serves its Senior Vice President, member airlines and their customers by: General Counsel and Secretary 2 • Assisting the airline industry in continuing to prov i d e James L. Casey the world’s safest system of transportation Vice President and • Transmitting technical expertise and operational Deputy General Counsel kn o w l e d g e among member airlines to improve safety, service and efficiency J. Donald Collier Vice President, • Advocating fair airline taxation and regulation world- Engineering, Maintenance and Materiel wide, ensuring a profitable and competitive industry Albert H. Prest Vice President, Operations Nestor N. Pylypec Vice President, Industry Services Michael D. Wascom Vice President, Communications Richard T. Brandenburg Treasurer and Chief Financial Officer David A. Swierenga Chief Economist RepLayout for final pdf 8/28/2001 9:24 AM Page 3 [ c u s t o m e r s ] Table of Contents Officers . .2 The member airlines of the Air Mission . .2 President’s Letter . .5 Transport Association are committed to Goals . .5 providing the highest level of customer Highlights . -

INTERNATIONAL CONFERENCE on AIR LAW (Montréal, 20 April to 2
DCCD Doc No. 28 28/4/09 (English only) INTERNATIONAL CONFERENCE ON AIR LAW (Montréal, 20 April to 2 May 2009) CONVENTION ON COMPENSATION FOR DAMAGE CAUSED BY AIRCRAFT TO THIRD PARTIES AND CONVENTION ON COMPENSATION FOR DAMAGE TO THIRD PARTIES, RESULTING FROM ACTS OF UNLAWFUL INTERFERENCE INVOLVING AIRCRAFT (Presented by the Air Crash Victims Families Group) 1. INTRODUCTION – SUPPLEMENTAL AND OTHER COMPENSATIONS 1.1 The apocalyptic terrorist attack by the means of four hi-jacked planes committed against the World Trade Center in New York, NY , the Pentagon in Arlington, VA and the aborted flight ending in a crash in the rural area in Shankville, PA ON September 11th, 2001 is the only real time example that triggered this proposed Convention on Compensation for Damage to Third Parties from Acts of Unlawful Interference Involving Aircraft. 1.2 It is therefore important to look towards the post incident resolution of this tragedy in order to adequately and pro actively complete ONE new General Risk Convention (including compensation for ALL catastrophic damages) for the twenty first century. 2. DISCUSSION 2.1 Immediately after September 11th, 2001 – the Government and Congress met with all affected and interested parties resulting in the “Air Transportation Safety and System Stabilization Act” (Public Law 107-42-Sept. 22,2001). 2.2 This Law provided the basis for Rules and Regulations for: a) Airline Stabilization; b) Aviation Insurance; c) Tax Provisions; d) Victims Compensation; and e) Air Transportation Safety. DCCD Doc No. 28 - 2 - 2.3 The Airline Stabilization Act created the legislative vehicle needed to reimburse the air transport industry for their losses of income as a result of the flight interruption due to the 911 attack. -

Global Volatility Steadies the Climb
WORLD AIRLINER CENSUS Global volatility steadies the climb Cirium Fleet Forecast’s latest outlook sees heady growth settling down to trend levels, with economic slowdown, rising oil prices and production rate challenges as factors Narrowbodies including A321neo will dominate deliveries over 2019-2038 Airbus DAN THISDELL & CHRIS SEYMOUR LONDON commercial jets and turboprops across most spiking above $100/barrel in mid-2014, the sectors has come down from a run of heady Brent Crude benchmark declined rapidly to a nybody who has been watching growth years, slowdown in this context should January 2016 low in the mid-$30s; the subse- the news for the past year cannot be read as a return to longer-term averages. In quent upturn peaked in the $80s a year ago. have missed some recurring head- other words, in commercial aviation, slow- Following a long dip during the second half Alines. In no particular order: US- down is still a long way from downturn. of 2018, oil has this year recovered to the China trade war, potential US-Iran hot war, And, Cirium observes, “a slowdown in high-$60s prevailing in July. US-Mexico trade tension, US-Europe trade growth rates should not be a surprise”. Eco- tension, interest rates rising, Chinese growth nomic indicators are showing “consistent de- RECESSION WORRIES stumbling, Europe facing populist backlash, cline” in all major regions, and the World What comes next is anybody’s guess, but it is longest economic recovery in history, US- Trade Organization’s global trade outlook is at worth noting that the sharp drop in prices that Canada commerce friction, bond and equity its weakest since 2010. -

Comprehensive Plan Transportation-123 2015-2035 • Low – 459,617
Chapter 5 Transportation Introduction This Transportation Element (TE) is prepared in accordance with the GMA. Contained within the TE are projects and implementation measures necessary to effectively serve planned land use throughout unincorporated Clark County. Importantly, this element provides guidance for the design, construction and operation of transportation facilities and services through the year 2035. Purpose and Background The purpose of the TE is to present a plan for transportation facilities and services needed to support the county’s 2015-2035 future land use map. The TE recommends specific arterial roadway projects for the unincorporated county in order to meet roadway safety and capacity needs. However, it also recommends various implementation strategies to guide the county in its participation in regional transportation planning. Implementation strategies provide guidance on such issues as: • land use-transportation concurrency; • arterial, highway and transit level-of-service; • transit emphasis corridors; • access management; • transportation demand management (TDM); • non-motorized transportation; • air quality conformance; and • freight and goods mobility. The county’s TE provides an estimate of expenditures and revenues associated with implementing various recommended transportation improvements. It also recommends a financial strategy that would ensure needed transportation improvements are funded. It should be noted that the transportation element can be amended and supplemented by special studies that later provide more detailed policy direction and project recommendations. These special studies would maintain consistency with the countywide transportation element, while also qualifying and refining its recommendations. Description of Historical Growth and Development Clark County’s population was estimated at 448,500 in 2015 making it the 5th most populous county in Washington State. -

Actions Needed to Improve Airline Customer Service and Minimize Long, On-Board Delays
Before the Committee on Transportation and Infrastructure Subcommittee on Aviation United States House of Representatives For Release on Delivery Expected at 2:00 p.m. EDT Actions Needed To Wednesday September 26, 2007 Improve Airline CC-2007-099 Customer Service and Minimize Long, On-Board Delays Statement of The Honorable Calvin L. Scovel III Inspector General U.S. Department of Transportation Mr. Chairman and Members of the Subcommittee: We are pleased to be here today to discuss airline customer service issues and the actions needed from the Department of Transportation (DOT), Federal Aviation Administration (FAA), airlines, and airports to minimize long, on-board delays. This hearing is both timely and important given the record-breaking flight delays, cancellations, diversions, and on-board tarmac delays that air travelers have already experienced this year. Based on the first 7 months of the year: • Nearly 28 percent of flights were delayed, cancelled, or diverted—with airlines’ on-time performance at the lowest percentage (72 percent) recorded in the last 10 years. • Not only are there more delays, but also longer delay periods. Of those flights arriving late, passengers experienced a record-breaking average flight arrival delay of 57 minutes, up nearly 3 minutes from 2006. • More than 54,000 flights affecting nearly 3.7 million passengers experienced taxi-in and taxi-out times of 1 to 5 hours or more. This is an increase of nearly 42 percent as compared to the same period in 2006. As you know, Secretary Peters has expressed serious concerns about the airlines’ treatment of passengers during extended ground delays. -

Laguardia Airport
Aviation Department Traffic Statistics: D.Wilson, J. Cuneo THE PORT AUTHORITY OF NY & NJ JUNE 2007 TRAFFIC REPORT Current month,12 months ending,year-to-date totals Showing percentage change from prior year period Month Year-to-date 12 Months Ending LGA Current % Current % Current % PASSENGERS Domestic 2,094,634 -5.1 11,718,462 -4.2 23,986,727 -2.1 International 109,583 -5.8 589,898 -6.6 1,272,111 -6.2 Total Revenue Passengers 2,204,217 -5.1 12,308,360 -4.3 25,258,838 -2.3 Non Revenue Passengers 64,627 -7.7 341,790 -10.8 718,978 -10.9 Note: Commuter - Regional Pax incl. in above 463,366 0.7 2,500,200 4.6 5,085,284 7.0 FLIGHTS Domestic 28,253 -5.9 177,628 -1.6 360,527 -0.6 International 1,768 -6.5 10,442 -4.5 21,483 -3.7 General Aviation 1,208 2.0 7,322 -1.0 14,318 -3.1 Total 31,229 -5.7 195,392 -1.8 396,328 -0.8 Note: Commuter - Regional Flights incl. in above 14,256 -3.4 91,055 3.0 184,743 4.2 FREIGHT (in short tons) Domestic 709 -46.0 4,948 -36.0 10,966 -31.6 International 16 -20.0 114 -13.0 229 -29.3 Total 725 -45.6 5,062 -35.6 11,195 -31.6 MAIL (in short tons) Total 137 -79.5 1,020 -78.7 1,956 -79.0 Ground Transportation Paid Parked Cars 168,554 -10.5 937,956 -11.7 1,967,853 -9.4 Ground Transpo.Counter Passengers 11,405 -19.1 65,725 -22.5 132,761 -13.4 Airport Coach Passengers 37,000 -31.1 186,174 -10.5 390,968 -6.6 Taxis Dispatched 324,894 -2.0 1,898,376 2.3 3,769,281 -0.2 Air Transport Association Carriers (USA) Passengers:Domestic Enplaned (000) 44,217 2.3 243,391 1.2 486,615 1.5 Passengers:International Enplaned (000) 6,384 2.3