Flyxcdb—A Resource for Drosophila Cell Surface and Secreted Proteins and Their Extracellular Domains
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Figure S1 Functional Characterisation of Snmp:GFP
doi: 10.1038/nature06328 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE LEGENDS Figure S1 | Functional characterisation of SNMP fusion proteins. Dose-response curve for cVA in Or67d neurons of wild-type (Berlin), SNMP mutant (Or67d- GAL4/+;SNMP1/SNMP2), SNMP:GFP rescue (Or67d-GAL4/UAS- SNMP:GFP;SNMP1/SNMP2) and YFP(1):Or83b/SNMP:YFP(2) rescue (Or67d:GAL4,UAS-YFP(1):Or83b/UAS-SNMP:YFP(2);SNMP1,Or83b2/SNMP2,Or83b1 ) animals. Mean responses are plotted (± s.e.m; wild-type n=47, SNMP mutant n=46, SNMP:GFP rescue n=20; YFP(1):Or83b/SNMP:YFP(2) rescue n=22; ≤4 sensilla/animal, mixed genders). Wild-type and SNMP:GFP rescue responses to cVA are not significantly different (ANOVA; p>0.1175). YFP(1):Or83b/SNMP:YFP(2) rescue responses to cVA are highly significantly different from SNMP mutants and from wild- type (ANOVA; p<0.0001), indicating partial rescue. Figure S2 | Cell type-specific rescue of SNMP expression. a, Immunostaining for mCD8:GFP (anti-GFP, green) and LUSH (magenta) in LUSH-GAL4/UAS-mCD8:GFP animals reveals faithful recapitulation of endogenous expression by the LUSH-GAL4 driver. b, Two-colour RNA in situ hybridisation for SNMP (green) and Or67d (magenta) in antennal sections of wild-type, Or67d neuron SNMP rescue (Or67d-GAL4/UAS- SNMP;SNMP1/SNMP2) and support cell SNMP rescue (LUSH-GAL4/UAS- SNMP;SNMP1/SNMP2) animals. www.nature.com/nature 1 Benton et al., Figure S1 ) -1 wild-type 120 SNMP:GFP rescue 80 YFP(1):Or83b/SNMP:YFP(2) rescue 40 Corrected response (spikes s 0 SNMP-/- 0 0.1 1 10 100 cVA (%) www.nature.com/nature 2 Benton -

Supp Table 6.Pdf
Supplementary Table 6. Processes associated to the 2037 SCL candidate target genes ID Symbol Entrez Gene Name Process NM_178114 AMIGO2 adhesion molecule with Ig-like domain 2 adhesion NM_033474 ARVCF armadillo repeat gene deletes in velocardiofacial syndrome adhesion NM_027060 BTBD9 BTB (POZ) domain containing 9 adhesion NM_001039149 CD226 CD226 molecule adhesion NM_010581 CD47 CD47 molecule adhesion NM_023370 CDH23 cadherin-like 23 adhesion NM_207298 CERCAM cerebral endothelial cell adhesion molecule adhesion NM_021719 CLDN15 claudin 15 adhesion NM_009902 CLDN3 claudin 3 adhesion NM_008779 CNTN3 contactin 3 (plasmacytoma associated) adhesion NM_015734 COL5A1 collagen, type V, alpha 1 adhesion NM_007803 CTTN cortactin adhesion NM_009142 CX3CL1 chemokine (C-X3-C motif) ligand 1 adhesion NM_031174 DSCAM Down syndrome cell adhesion molecule adhesion NM_145158 EMILIN2 elastin microfibril interfacer 2 adhesion NM_001081286 FAT1 FAT tumor suppressor homolog 1 (Drosophila) adhesion NM_001080814 FAT3 FAT tumor suppressor homolog 3 (Drosophila) adhesion NM_153795 FERMT3 fermitin family homolog 3 (Drosophila) adhesion NM_010494 ICAM2 intercellular adhesion molecule 2 adhesion NM_023892 ICAM4 (includes EG:3386) intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)adhesion NM_001001979 MEGF10 multiple EGF-like-domains 10 adhesion NM_172522 MEGF11 multiple EGF-like-domains 11 adhesion NM_010739 MUC13 mucin 13, cell surface associated adhesion NM_013610 NINJ1 ninjurin 1 adhesion NM_016718 NINJ2 ninjurin 2 adhesion NM_172932 NLGN3 neuroligin -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -
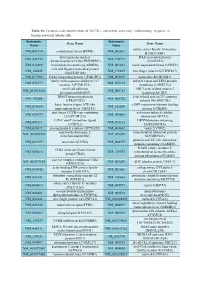
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Sharmin Supple Legend 150706
Supplemental data Supplementary Figure 1 Generation of NPHS1-GFP iPS cells (A) TALEN activity tested in HEK 293 cells. The targeted region was PCR-amplified and cloned. Deletions in the NPHS1 locus were detected in four clones out of 10 that were sequenced. (B) PCR screening of human iPS cell homologous recombinants (C) Southern blot screening of human iPS cell homologous recombinants Supplementary Figure 2 Human glomeruli generated from NPHS1-GFP iPS cells (A) Morphological changes of GFP-positive glomeruli during differentiation in vitro. A different aggregate from the one shown in Figure 2 is presented. Lower panels: higher magnification of the areas marked by rectangles in the upper panels. Note the shape changes of the glomerulus (arrowheads). Scale bars: 500 µm. (B) Some, but not all, of the Bowman’s capsule cells were positive for nephrin (48E11 antibody: magenta) and GFP (green). Scale bars: 10 µm. Supplementary Figure 3 Histology of human podocytes generated in vitro (A) Transmission electron microscopy of the foot processes. Lower magnification of Figure 4H. Scale bars: 500 nm. (B) (C) The slit diaphragm between the foot processes. Higher magnification of the 1 regions marked by rectangles in panel A. Scale bar: 100 nm. (D) Absence of mesangial or vascular endothelial cells in the induced glomeruli. Anti-PDGFRβ and CD31 antibodies were used to detect the two lineages, respectively, and no positive signals were observed in the glomeruli. Podocytes are positive for WT1. Nuclei are counterstained with Nuclear Fast Red. Scale bars: 20 µm. Supplementary Figure 4 Cluster analysis of gene expression in various human tissues (A) Unbiased cluster analysis across various human tissues using the top 300 genes enriched in GFP-positive podocytes. -

Figure S1. GO Analysis of Genes in Glioblastoma Cases That Showed Positive and Negative Correlations with TCIRG1 in the GSE16011 Cohort
Figure S1. GO analysis of genes in glioblastoma cases that showed positive and negative correlations with TCIRG1 in the GSE16011 cohort. (A‑C) GO‑BP, GO‑CC and GO‑MF terms of genes that showed positive correlations with TCIRG1, respec‑ tively. (D‑F) GO‑BP, GO‑CC and GO‑MF terms of genes that showed negative correlations with TCIRG1. Red nodes represent gene counts, and black bars represent negative 1og10 P‑values. TCIRG1, T cell immune regulator 1; GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function. Table SI. Genes correlated with T cell immune regulator 1. Gene Name Pearson's r ARPC1B Actin‑related protein 2/3 complex subunit 1B 0.756 IL4R Interleukin 4 receptor 0.695 PLAUR Plasminogen activator, urokinase receptor 0.693 IFI30 IFI30, lysosomal thiol reductase 0.675 TNFAIP3 TNF α‑induced protein 3 0.675 RBM47 RNA binding motif protein 47 0.666 TYMP Thymidine phosphorylase 0.665 CEBPB CCAAT/enhancer binding protein β 0.663 MVP Major vault protein 0.660 BCL3 B‑cell CLL/lymphoma 3 0.657 LILRB3 Leukocyte immunoglobulin‑like receptor B3 0.656 ELF4 E74 like ETS transcription factor 4 0.652 ITGA5 Integrin subunit α 5 0.651 SLAMF8 SLAM family member 8 0.647 PTPN6 Protein tyrosine phosphatase, non‑receptor type 6 0.641 RAB27A RAB27A, member RAS oncogene family 0.64 S100A11 S100 calcium binding protein A11 0.639 CAST Calpastatin 0.638 EHBP1L1 EH domain‑binding protein 1‑like 1 0.638 LILRB2 Leukocyte immunoglobulin‑like receptor B2 0.629 ALDH3B1 Aldehyde dehydrogenase 3 family member B1 0.626 GNA15 G protein -

Mapping Transmembrane Binding Partners for E-Cadherin Ectodomains
SUPPLEMENTARY INFORMATION TITLE: Mapping transmembrane binding partners for E-cadherin ectodomains. AUTHORS: Omer Shafraz 1, Bin Xie 2, Soichiro Yamada 1, Sanjeevi Sivasankar 1, 2, * AFFILIATION: 1 Department of Biomedical Engineering, 2 Biophysics Graduate Group, University of California, Davis, CA 95616. *CORRESPONDING AUTHOR: Sanjeevi Sivasankar, Tel: (530)-754-0840, Email: [email protected] Figure S1: Western blots a. EC-BioID, WT and Ecad-KO cell lysates stained for Ecad and tubulin. b. HRP-streptavidin staining of biotinylated proteins eluted from streptavidin coated magnetic beads incubated with cell lysates of EC-BioID with (+) and without (-) exogenous biotin. c. C-BioID, WT and Ecad-KO cell lysates stained for Ecad and tubulin. d. HRP-streptavidin staining of biotinylated proteins eluted from streptavidin coated magnetic beads incubated with cell lysates of C-BioID with (+) and without (-) exogenous biotin. (+) Biotin (-) Biotin Sample 1 Sample 2 Sample 3 Sample 4 Sample 1 Sample 2 Sample 3 Sample 4 Percent Percent Percent Percent Percent Percent Percent Percent Gene ID Coverage Coverage Coverage Coverage Coverage Coverage Coverage Coverage CDH1 29.6 31.4 41.1 36.5 10.8 6.7 28.8 29.1 DSG2 26 14.6 45 37 0.8 1.9 1.6 18.7 CXADR 30.2 26.2 32.7 27.1 0.0 0.0 0.0 6.9 EFNB1 24.3 30.6 24 30.3 0.0 0.0 0.0 0.0 ITGA2 16.5 22.2 30.1 33.4 1.1 1.1 5.2 7.2 CDH3 21.8 9.7 20.6 25.3 1.3 1.3 0.0 0.0 ITGB1 11.8 16.7 23.9 20.3 0.0 2.9 8.5 5.8 DSC3 9.7 7.5 11.5 13.3 0.0 0.0 2.6 0.0 EPHA2 23.2 31.6 31.6 30.5 0.8 0.0 0.0 5.7 ITGB4 21.8 27.8 33.1 30.7 0.0 1.2 3.9 4.4 ITGB3 23.5 22.2 26.8 24.7 0.0 0.0 5.2 9.1 CDH6 22.8 18.1 28.6 24.3 0.0 0.0 0.0 9.1 CDH17 8.8 12.4 20.7 18.4 0.0 0.0 0.0 0.0 ITGB6 12.7 10.4 14 17.1 0.0 0.0 0.0 1.7 EPHB4 11.4 8.1 14.2 16.3 0.0 0.0 0.0 0.0 ITGB8 5 10 15 17.6 0.0 0.0 0.0 0.0 ITGB5 6.2 9.5 15.2 13.8 0.0 0.0 0.0 0.0 EPHB2 8.5 4.8 9.8 12.1 0.0 0.0 0.0 0.0 CDH24 5.9 7.2 8.3 9 0.0 0.0 0.0 0.0 Table S1: EC-BioID transmembrane protein hits. -

Phosphotyrosine Recognition Domains: the Typical, the Atypical and the Versatile Tomonori Kaneko1†, Rakesh Joshi1†, Stephan M Feller2 and Shawn SC Li1*
Kaneko et al. Cell Communication and Signaling 2012, 10:32 http://www.biosignaling.com/content/10/1/32 REVIEW Open Access Phosphotyrosine recognition domains: the typical, the atypical and the versatile Tomonori Kaneko1†, Rakesh Joshi1†, Stephan M Feller2 and Shawn SC Li1* Abstract SH2 domains are long known prominent players in the field of phosphotyrosine recognition within signaling protein networks. However, over the years they have been joined by an increasing number of other protein domain families that can, at least with some of their members, also recognise pTyr residues in a sequence-specific context. This superfamily of pTyr recognition modules, which includes substantial fractions of the PTB domains, as well as much smaller, or even single member fractions like the HYB domain, the PKCδ and PKCθ C2 domains and RKIP, represents a fascinating, medically relevant and hence intensely studied part of the cellular signaling architecture of metazoans. Protein tyrosine phosphorylation clearly serves a plethora of functions and pTyr recognition domains are used in a similarly wide range of interaction modes, which encompass, for example, partner protein switching, tandem recognition functionalities and the interaction with catalytically active protein domains. If looked upon closely enough, virtually no pTyr recognition and regulation event is an exact mirror image of another one in the same cell. Thus, the more we learn about the biology and ultrastructural details of pTyr recognition domains, the more does it become apparent that nature cleverly combines and varies a few basic principles to generate a sheer endless number of sophisticated and highly effective recognition/regulation events that are, under normal conditions, elegantly orchestrated in time and space. -
Supplementary Table 2
Table S2 Potential targets of miR-9-3 in MCF-7 cells Mean of Intensities Ratio Gene Symbol Human Gene Description Ctrl miR-9 miR-9* miR-9/Ctrl miR-9*/Ctrl FBXW2 F-box and WD repeat domain containing 2 109.61 502.26 513.97 4.58 4.69 FAM62A family with sequence similarity 62, member A 65.28 287.19 245.59 4.40 3.76 SMEK1 SMEK homolog 1, suppressor of mek1 (Dictyostelium) 87.12 333.06 343.35 3.82 3.94 KLHDC5 kelch domain containing 5 99.92 344.30 420.00 3.45 4.20 CC2D1B coiled-coil and C2 domain containing 1B 296.83 1013.51 988.06 3.41 3.33 GPAM glycerol-3-phosphate acyltransferase, mitochondrial 87.09 286.46 278.41 3.29 3.20 CUL4A cullin 4A 47.99 155.16 200.94 3.23 4.19 SMC1A structural maintenance of chromosomes 1A 126.43 403.56 465.37 3.19 3.68 CACNB2 calcium channel, voltage-dependent, beta 2 subunit 87.11 266.88 279.56 3.06 3.21 SLC6A6 solute carrier family 6, member 6 137.27 414.29 447.48 3.02 3.26 SEC31A SEC31 homolog A (S. cerevisiae) 79.11 234.96 280.71 2.97 3.55 PTBP2 polypyrimidine tract binding protein 2 495.58 1437.74 1282.95 2.90 2.59 C3orf58 chromosome 3 open reading frame 58 62.72 175.93 152.28 2.80 2.43 HIC2 hypermethylated in cancer 2 157.64 427.84 447.94 2.71 2.84 KIAA0562 KIAA0562 73.31 195.78 213.59 2.67 2.91 LPP LIM domain containing preferred translocation partner in lipoma 60.87 161.90 184.81 2.66 3.04 FNIP1 folliculin interacting protein 1 161.56 421.82 464.73 2.61 2.88 TFRC transferrin receptor (p90, CD71) 95.11 244.87 284.62 2.57 2.99 RNF111 ring finger protein 111 44.75 112.69 131.59 2.52 2.94 USP31 ubiquitin specific -

Differential Transcriptome Profile of Peripheral White Cells to Identify Biomarkers Involved in Oxaliplatin Induced Neuropathy
Supplementary Material for Differential Transcriptome Profile of Peripheral White Cells to Identify Biomarkers Involved in Oxaliplatin Induced Neuropathy Fold Gene p_value Description Change Genes whose expression increased after three cycles of oxaliplatin therapy (in alphabetical order) A4GALT 5.8 0.017 alpha 1,4-galactosyltransferase [Source:HGNC Symbol;Acc:18149] AGRN 2.8 0.001 agrin [Source:HGNC Symbol;Acc:329] AIF1 3.4 0.000 allograft inflammatory factor 1 [Source:HGNC Symbol;Acc:352] AKAP12 4.5 0.007 A kinase (PRKA) anchor protein 12 [Source:HGNC Symbol;Acc:370] AKR1C2 8.0 0.006 aldo-keto reductase family 1, member C2 [Source:HGNC Symbol;Acc:385] ALDH3A1 4.3 0.042 aldehyde dehydrogenase 3 family, member A1 [Source:HGNC Symbol;Acc:405] AMOTL2 3.5 0.047 angiomotin like 2 [Source:HGNC Symbol;Acc:17812] ANTXR1 4.5 0.006 anthrax toxin receptor 1 [Source:HGNC Symbol;Acc:21014] ANXA1 1.7 0.031 annexin A1 [Source:HGNC Symbol;Acc:533] ANXA2 1.8 0.027 annexin A2 [Source:HGNC Symbol;Acc:537] AP1M2 3.8 0.026 adaptor-related protein complex 1, mu 2 subunit [Source:HGNC Symbol;Acc:558] AP2S1 1.7 0.033 adaptor-related protein complex 2, sigma 1 subunit [Source:HGNC Symbol;Acc:565] ARHGEF17 5.0 0.026 Rho guanine nucleotide exchange factor (GEF) 17 [Source:HGNC Symbol;Acc:21726] ASGR1 2.9 0.012 asialoglycoprotein receptor 1 [Source:HGNC Symbol;Acc:742] ASPHD1 7.2 0.028 aspartate beta-hydroxylase domain containing 1 [Source:HGNC Symbol;Acc:27380] ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 [Source:HGNC ATP5EP2 5.2 0.003 Symbol;Acc:34026] ATP5F1 2.3 0.017 ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 [Source:HGNC Symbol;Acc:840] ATP5I 3.5 0.001 ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E [Source:HGNC Symbol;Acc:846] ATPIF1 2.0 0.030 ATPase inhibitory factor 1 [Source:HGNC Symbol;Acc:871] J. -

High Resolution Genome Wide Expression Analysis of Single Myofibers Using SMART-Seq
bioRxiv preprint doi: https://doi.org/10.1101/724393; this version posted October 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Blackburn et al. 2019 Manuscript High Resolution Genome Wide Expression Analysis of Single Myofibers Using SMART-Seq Darren M. Blackburn1,2,*, Felicia Lazure1,2,*, Aldo H. Corchado1, Theodore J. Perkins3,4, Hamed S. Najafabadi1, Vahab D. Soleimani1,2,5 1Department of Human Genetics, McGill University, 3640 rue University, Montreal, QC, H3A 0C7 Canada 2Molecular and Regenerative Medicine Axis, Lady Davis Institute for Medical ResearcH, JewisH General Hospital, 3755 Chemin de la Cote-Sainte-CatHerine, Montreal, QC, H3T 1E2, Canada 3Sprott Center for Stem Cell Research, Ottawa Hospital Research Institute, 501 Smyth Road, Ottawa, ON, K1H 8L6, Canada 4Department of BiocHemistry, Microbiology and Immunology, University of Ottawa, 451 SmytH Road, Ottawa, ON, K1H 8M5, Canada 5To whom correspondence sHould be addressed: VaHab D. Soleimani: Department of Human Genetics, McGill University, 3640 rue University, Montreal, QC, H3A 0C7, Canada; [email protected]; TelepHone number: +1 514 340 8222 ext. 26136; Fax number: +1 514 340-7502. *These autHors made equal contributions Running title: Single myofiber RNA-Seq Key Words: Single Myofiber, Skeletal Muscle, SMART-Seq, Gene Expression Analysis, Myofiber Heterogeneity, Aging skeletal muscle Word count: 3930 1 bioRxiv preprint doi: https://doi.org/10.1101/724393; this version posted October 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved.