Gut Microbiota Prevents Sugar Alcohol-Induced Diarrhea
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

GRAS Notice 789 for Erythritol
GRAS Notice (GRN) No. 789 https://www.fda.gov/food/generally-recognized-safe-gras/gras-notice-inventory. Toi• Strategies ~~~~G~~[)) JUN 7 20'8 Innovative solutions Sound science OFFICE OF FOOD ADDITIVE SAFE1Y June 5, 2018 Dr. Dennis Keefe Director, Division of Biotechnology and GRAS Notice Review Office of Food Additive Safety (HFS-200) Center for Food Safety and Applied Nutrition Food and Drug Administration 5100 Paint Branch Parkway College Park, MD 20740-3835 Subject: GRAS Notification - Erythritol Dear Dr. Keefe: On behalf of Cargill, Incorporated, ToxStrategies, Inc. (its agent) is submitting, for FDA review, a copy of the GRAS notification as required. The enclosed document provides notice of a claim that the food ingredient, erythritol, described in the enclosed notification is exempt from the premarket approval requirement of the Federal Food, Drug, and Cosmetic Act because it has been determined to be generally recognized as safe (GRAS), based on scientific procedures, for addition to food. If you have any questions or require additional information, please do not hesitate to contact me at 630-352-0303, or [email protected]. Sincerely, (b) (6) Donald F. Schmitt, M.P.H. Senior Managing Scientist ToxStrategies, Inc., 931 W. 75th St. , Suite 137, PMB 263, Naperville, IL 60565 1 Office (630) 352-0303 • www.toxstrategies.com GRAS Determination of Erythritol for Use in Human Food JUNES,2018 Innovative solutions s ,..,.,',--.r-.r--.r--. OFFICE OF FOOD ADDITIVE SAFE1Y GRAS Determination of Erythritol for Use in Human Food SUBMITTED BY: Cargill, Incorporated 15407 McGinty Road West Wayzata, MN 55391 SUBMITTED TO: U.S. Food and Drug Administration Center for Food Safety and Applied Nutrition Office of Food Additive Safety HFS-200 5100 Paint Branch Parkway College Park MD 20740-3835 CONTACT FOR TECHNICAL OR OTIIER INFORMATION Donald F. -

Study of the Partial and Exhaustive Intramolecular Dehydration of D-Sorbitol
Available online at http://journal-of-agroalimentary.ro Journal of Journal of Agroalimentary Processes and Agroalimentary Processes and Technologies 2013, 19(2), 259-270 Technologies Study of the partial and exhaustive intramolecular dehydration of D-sorbitol Ileana Cocan *, Monica Viorica Negrea, Ionel Vasile Jianu ”Dunărea de Jos” University of Galati, Faculty of Food Science and Engineering, Domnească Street, 47, RO-800008, Galati, Romania Received: 07 February 2013; Accepted: 09 March 2013 ______________________________________________________________________________________ Abstract Sweetener, humectants, sequestrant, texturizer, stabilizer, bulking agent, D-sorbitol represents a leading product in the processing of the polysorbate class. The scope of this work is to obtain details on the preparation and accession of D-sorbitol mono- and dianhydride as a method of chemical protection of primary and secondary hydroxyl functional groups (1:4) (3:6) as part of the strategy of controlled processing of polysorbates containing “homogeneous” polyoxyethylenic chains (n = 3, 6, 9, 18). This work is investigating the dependence of yield of internal dehydration (partial and/or exhaustive, direct of D-sorbitol) on temperature (100-160°C and 120-200°C , respectively ) and duration (5–35 minutes and 10–100 minutes , respectively ) for the processing of 1,4-sorbitan and 2,5-isosorbide. The evolution of the hydroxyl number (mg KOH/g D-sorbitol) (mg KOH/g 1,4-sorbitan) related to the theoretical (initial) value was followed and optimal values for the yield and dehydration parameters were established. Also in this work, the mathematical modeling of the dependence curves experimentally registered is realized. Keywords : izosorbide, D-sorbitol, D-glucitol, D-sorbit, 1,4-sorbitan ______________________________________________________________________________________ 1. -
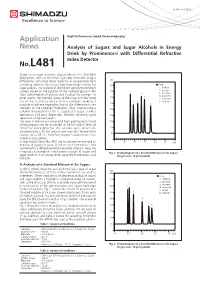
Analysis of Sugars and Sugar Alcohols in Energy Drink by Prominence-I with Differential Refreactive Index Detector
LAAN-A-LC-E258 Application High Performance Liquid Chromatography News Analysis of Sugars and Sugar Alcohols in Energy Drink by Prominence-i with Differential Refractive Index Detector No.L481 Sugars and sugar alcohols display almost no ultraviolet absorption, and are therefore typically detected using a differential refractive index detector or evaporative light uRI scattering detector. By using a ligand exchange column for 80 ■ Peaks sugar analysis, it is possible to distinguish among the different 1. maltose 70 2. glucose isomers based on the position of the hydroxyl group in the 1 4 3. fructose chair conformation of glucose and fructose for example. In 4. erythritol 60 5 other words, the hydroxyl group of the sugar and the metal 5. mannitol ion of the stationary phase form a complex, making it 2 3 6. sorbitol possible to achieve separation due to the difference in the 50 6 strength of the complex formation. Also, maintaining a 40 column temperature of 80 °C suppresses sugar anomer separation and peak dispersion, thereby achieving good 30 separation of adjacent peaks. The new Prominence-i integrated high-performance liquid 20 chromatograph can be connected to the RID-20A differential refractive index detector. The column oven, which can 10 accommodate a 30 cm column and maintain temperature 0 control up to 85 °C, therefore supports applications that require a long column. In Application News No. 467, we introduced an example of 0 5 10 15 20 25 min analysis of sugars in juice, in which the Prominence-i was connected to a differential refractive index detector. Here, we introduce an example of simultaneous analysis of sugars and Fig. -

Sorbitol Malabsorption 1/2
Internal Medicine and Gastroenterology Clinic Dres. Mares,M.Hanig,Mares, Hanig, Rambow, S.Blau, Blau, M.Seip, Hanig, Seip, A.Borchers,Blau,Kirchner, Hochstr. Hochstr. Hochstr. 43, 60313 43, 43, 60313 60313 Frankfurt/Main, Frankfurt/M., Frankfurt/M., www.gastroenterologie-ffm.de www.gastroenterologie-ffm.de www.gastroenterologie-ffm.de Nutrition hints for sorbitol malabsorption 1/2 Sorbitol intolerance Sorbitol incompatibility 1. What does this mean for nutrition? Avoid sorbitol as a sweetener and foods high in sorbitol. Small amount of sorbitol can often be tolerated (at most 10-10 g per day, sometimes less). In order to prevent general upper GI problems, easily digestible foods that do not cause gas are recommended. 2. Key points about foods – Avoid sorbitol and foods containing sorbitol – easily digestible food that does not cause gas 3. vegetables and fruits low in fibre (see 5) 4. Choice of foods The following foods are high in sorbitol and not suitable: – Sorbitol as sweetener: e.g. Sionon, Flarom, diabetic sweetener – Dietetic foods produced with sorbitol: for example, diabetic marmalades, diabetic sweets, diabetic baked goods – Types of fruit which have a naturally high sorbitol content: Apples, pears, cherries, prunes, plums, dates fruits with seeds, such as mirabelles, apricots, nectarines, and all fruit syrups made from these types of fruits – Types of fruit which have a naturally low sorbitol content: Berry fruits such as strawberries, raspberries, blackberries, blueberries, currants, gooseberries, citrus fruits, bananas, pineapples, kiwis – Sorbitol as a coating for: sultanas, raisins, and dried fruit or candied fruit – Sorbitol in sweets: chewing gum, jelly babies, jelly fruits, candies, chocolate bars, filled wafers, chocolate, etc. -

SAFETY DATA SHEET Isopropyl Alcohol
SAFETY DATA SHEET Isopropyl Alcohol Section 1. Identification GHS product identifier : Isopropyl Alcohol Chemical name : Isopropyl alcohol Other means of : isopropanol; 2-Propanol identification Product type : Liquid. Product use : Synthetic/Analytical chemistry. Synonym : isopropanol; 2-Propanol SDS # : 001105 Supplier's details : Airgas USA, LLC and its affiliates 259 North Radnor-Chester Road Suite 100 Radnor, PA 19087-5283 1-610-687-5253 24-hour telephone : 1-866-734-3438 Section 2. Hazards identification OSHA/HCS status : This material is considered hazardous by the OSHA Hazard Communication Standard (29 CFR 1910.1200). Classification of the : FLAMMABLE LIQUIDS - Category 2 substance or mixture EYE IRRITATION - Category 2A SPECIFIC TARGET ORGAN TOXICITY (SINGLE EXPOSURE) (Narcotic effects) - Category 3 GHS label elements Hazard pictograms : Signal word : Danger Hazard statements : May form explosive mixtures with air. Highly flammable liquid and vapor. Causes serious eye irritation. May cause drowsiness or dizziness. Precautionary statements General : Read label before use. Keep out of reach of children. If medical advice is needed, have product container or label at hand. Prevention : Wear protective gloves. Wear eye or face protection. Keep away from heat, hot surfaces, sparks, open flames and other ignition sources. No smoking. Use explosion- proof electrical, ventilating, lighting and all material-handling equipment. Use only non- sparking tools. Take precautionary measures against static discharge. Keep container tightly closed. Use only outdoors or in a well-ventilated area. Avoid breathing vapor. Wash hands thoroughly after handling. Response : IF INHALED: Remove person to fresh air and keep comfortable for breathing. Call a POISON CENTER or physician if you feel unwell. -

Copyrighted Material
Index Abbreviations receptor sites 202, 211 weak 122 amino acids, Table 500–1 muscarinic 413 weak, pH calculation 147–8 nucleic acids 551 nicotinic 413 Acid–base nucleotides, Table 551 as quaternary ammonium salt 202 catalysis, enzymes 516 peptides and proteins 504 Acetylcholinesterase equilibria 121 phosphates and diphosphates 277 enzyme mechanism 519–21 interactions, predicting 155–7 structural, Table 14 hydrolysis of acetylcholine 279 Acidic reagents, Table 157 ACE (angiotensin-converting enzyme) inhibitors 279 Acidity enzyme action 532 Acetyl-CoA carboxylase, in fatty acid acidity constant 122 inhibitors 532 biosynthesis 595 bond energy effects 125 Acetal Acetyl-CoA (acetyl coenzyme A) definition 121 in etoposide 233 as acylating agent 262 electronegativity effects 125 formation 229 carboxylation to malonyl-CoA 595, hybridization effects 128 polysaccharides as polyacetals 232 609 inductive effects 125–7 as protecting group 230 Claisen reaction 381 influence of electronic and structural Acetal and ketal enolate anions 373 features 125–34 cyclic, as protecting groups 481 in Krebs cycle 585 and leaving groups, Table 189 groups in sucrose 231 from β-oxidation of fatty acids 388 pKa values 122–5 Acetaldehyde, basicity 139 as thioester 262, 373 resonance / delocalization effects 129–34 Acetamide, basicity 139 Acetylene, bonding molecular orbitals 31 Acidity (compounds) Acetaminophen, see paracetamol Acetylenes, acidity 128 acetone 130 Acetoacetyl-CoA, biosynthesis from N-Acetylgalactosamine, in blood group acetonitrile 365 acetyl-CoA 392 -

United States Patent Office
United States Patentaw Office ... ber of ether groupings on each molecule. These mix tures, however, if they analyze as containing an average 2,978,421. number of ether groups per molecule greater than one, NITRILE COPOLYMERS AND METHOD OF are capable of producing the insoluble nitrile polymers PREPARING SAME of this invention. Since the efficiency of the polyether cross-linking agent increases with the number of poten John A. Holloway, Cleveland, Ohio, assignor to The tially polymerizable groups on the molecule, it is much B. F. Goodrich Company, New York, N.Y., a corpo preferred to utilize polyethers containing an average of ration of New York - two or more alkenyl ether groupings per molecule. The No Drawing. Filed June 9, 1958, Ser. No. 740,566. 10 polyvinyl polyethers of the polyhydric alcohols within the above broad class are produced by reacting acetylene 9 Claims. (C. 260-17.4) with the polyhydric alcohol (or an alcoholate thereof) in a Reppe-type vinylation synthesis. The polycrotyl ethers of the polyhydric alcohols are also useful although they This invention relates to cross-linked alpha-beta ole 15 do not contain a terminal CHCC group. - finically unsaturated nitrile polymers and more particu Illustrative polyhydric alcohols of the above-described larly pertains to copolymers of alpha-beta olefinically class that may be utilized in the preparation of the poly. unsaturated nitriles and polyalkenyl polyethers of poly alkenyl polyether, cross-linking agent include the butane hydric alcohols and methods for their preparation. triols such as 1,2,3-butane triol, 2,3,4-trihydroxybutyric An object of this invention is the provision of insoluble, 20 acid, the aldotetroses such as erythrose and threose, keto cross-linked polynitriles which are capable of thickening tetroses such as erythrulose; the aldopentoses such as certain non-aqueous polar solvents. -

Polyols Have a Variety of Functional Properties That Make Them Useful Alternatives to Sugars in Applications Including Baked Goods
Polyols have a variety of functional properties that make them useful alternatives to sugars in applications including baked goods. Photo © iStockphoto.com/Synergee pg 22 09.12 • www.ift.org BY LYN NABORS and THERESA HEDRICK SUGAR REDUCTION WITH Polyols Polyols are in a unique position to assist with reduced-sugar or sugar-free reformulations since they can reduce calories and complement sugar’s functionality. ugar reduction will be an important goal over the of the product’s original characteristics may still be main- next few years as consumers, government, and in- tained with the replacement of those sugars by polyols. Sdustry alike have expressed interest in lower-calorie In addition, excellent, good-tasting sugar-free products and lower-sugar foods. The 2010 Dietary Guidelines for can be developed by using polyols. Polyols are in a unique Americans put a strong emphasis on consuming fewer position to assist with reduced-sugar or sugar-free refor- calories and reducing intake of added sugars. The In- mulations; since they are only partially digested and ab- stitute of Medicine (IOM) held a public workshop in sorbed, they can reduce calories and complement sugar’s November 2010 to discuss ways the food industry can functionality. Polyols provide the same bulk as sugars and use contemporary and innovative food processing tech- other carbohydrates. Additionally, polyols have a clean, nologies to reduce calorie intake in an effort to reduce sweet taste, which is important since consumers are not and prevent obesity, and in October 2011 recommended likely to sacrifice taste for perceived health benefits. Poly- front-of-package labeling that includes rating the product ols have a host of other functional properties that make based on added sugars content. -

N-BUTYL ALCOHOL
Right to Know Hazardous Substance Fact Sheet Common Name: n-BUTYL ALCOHOL Synonyms: Propyl Carbinol; n-Butanol CAS Number: 71-36-3 Chemical Name: 1-Butanol RTK Substance Number: 1330 Date: November 1998 Revision: January 2008 DOT Number: UN 1120 Description and Use EMERGENCY RESPONDERS >>>> SEE BACK PAGE n-Butyl Alcohol is a colorless liquid with a strong, sweet Hazard Summary alcohol odor. It is used as a solvent for fats, waxes, shellacs, Hazard Rating NJDOH NFPA resins, gums, and varnish, in making hydraulic fluids, and in HEALTH - 2 medications for animals. FLAMMABILITY - 3 REACTIVITY - 0 f ODOR THRESHOLD = 1 to 15 ppm FLAMMABLE f Odor thresholds vary greatly. Do not rely on odor alone to POISONOUS GASES ARE PRODUCED IN FIRE determine potentially hazardous exposures. CONTAINERS MAY EXPLODE IN FIRE Hazard Rating Key: 0=minimal; 1=slight; 2=moderate; 3=serious; Reasons for Citation 4=severe f n-Butyl Alcohol is on the Right to Know Hazardous f n-Butyl Alcohol can affect you when inhaled and by Substance List because it is cited by OSHA, ACGIH, DOT, passing through the skin. NIOSH, DEP, IRIS, NFPA and EPA. f Contact can irritate and burn the skin. f This chemical is on the Special Health Hazard Substance f n-Butyl Alcohol can irritate and burn the eyes with possible List. eye damage. f Inhaling n-Butyl Alcohol can irritate the nose, throat and lungs. f Exposure to n-Butyl Alcohol can cause headache, dizziness, nausea and vomiting. SEE GLOSSARY ON PAGE 5. f n-Butyl Alcohol can damage the liver, kidneys, hearing, and sense of balance. -

Williamson Ether Synthesis the Williamson Ether Synthesis Is an Organic Reaction, Forming an Ether from an Alkyl Halide and an Alcohol
The Williamson ether synthesis The Williamson ether synthesis is an organic reaction, forming an ether from an alkyl halide and an alcohol. This reaction was developed by Alexander Williamson in 1850. It involves the reaction of an alkoxide ion with a primary alkyl halide via an SN2 reaction. The Williamson reaction is widely used in both laboratory and industrial synthesis, and remains the simplest and most popular method of preparing ethers. Both symmetrical and asymmetrical ethers are easily prepared. The reaction for this week: an example of a Williamson ether synthesis acetaminophen ethyl iodide phenacetin starting material reagent product Phenacetin may be synthesized as an example of the Williamson ether synthesis The first synthesis of phenacetin was reported in 1878 by Harmon Morse. Procedure 1. Weigh an Extra-Strength Tylenol tablet. Pulverize the tablet with mortar and pestle. Weigh out 0.22 g and place it in a dry 15-ml round-bottom flask along with 0.28 g of finely pulverized K2CO3 (mortar and pestle) and 3.0 mL of butanone. Carefully add 0.28 mL of ethyl iodide with a syringe. 2. Add a stir bar; attach a microscale water-cooled condenser to the flask. Heat the mixture under reflux directly on a hot plate at medium setting for 1 hour. In the meantime, obtain the IR of acetaminophen. 3. Turn off the heat. Allow the mixture to cool down. Add 4 mL of water to the flask and transfer its contents to a 16 x 125 mm test tube with a screw cap. Rinse round-bottom flask 4 times with 1 mL of tert-butyl methyl ether (BME) and add the rinsings to the test tube. -

LIQUID-VAPOR EQUILIBRIUM THERMODYNAMICS of the FUSEL OIL: a CASE STUDY Jéssyka Jennifer Miranda Corrêa¹, Edilailsa Januário
LIQUID-VAPOR EQUILIBRIUM THERMODYNAMICS OF THE FUSEL OIL: A CASE STUDY Jéssyka Jennifer Miranda Corrêa¹, Edilailsa Januário de Melo², José Izaquiel Santos da Silva³ ¹Graduating in Chemical Engineering from the Federal University of the Jequitinhonha and Mucuri Valleys, Diamantina - MG, Brazil. ²PhD student in Chemical Engineering, College of Chemical Engineering - State University of Campinas - SP, Brazill. ³Professor at the Federal University of the Jequitinhonha and Mucuri Valleys, Diamantina - MG, Brazil. E-mail for contact: [email protected] Recebido em: 06/04/2019 – Aprovado em: 10/06/2019 – Publicado em: 30/06/2019 DOI: 10.18677/EnciBio_2019A183 ABSTRACT Fusel oil is a by-product of the process of recovering hydrated ethanol, consisting of a mixture of superior alcohols, ethanol, water, among other components. Its commercial interest is mainly due to the presence of isoamyl alcohol, one of the raw materials used in the synthesis of esters, which compounds are relevant for the chemical industry. The recovery of this superior alcohol involves unit operations based on phase equilibrium, which for multicomponent systems can be studied in software that simulates the operation of equipment, in order to present predictions of interactions between its components, reducing errors, time and costs of design. In the industrial, academic and scientific fields, there are numerous challenges regarding the design of separation processes and the monitoring of mixing effects in typical operations, and the Aspen Plus process simulation and optimization platform excels in solving these issues. The current study aims to predict the Liquid-Vapor Equilibrium of the fusel oil, which was considered as a mixture of five components, from the behavioral analysis of each of the binaries that compose it. -

How Is Alcohol Metabolized by the Body?
Overview: How Is Alcohol Metabolized by the Body? Samir Zakhari, Ph.D. Alcohol is eliminated from the body by various metabolic mechanisms. The primary enzymes involved are aldehyde dehydrogenase (ALDH), alcohol dehydrogenase (ADH), cytochrome P450 (CYP2E1), and catalase. Variations in the genes for these enzymes have been found to influence alcohol consumption, alcohol-related tissue damage, and alcohol dependence. The consequences of alcohol metabolism include oxygen deficits (i.e., hypoxia) in the liver; interaction between alcohol metabolism byproducts and other cell components, resulting in the formation of harmful compounds (i.e., adducts); formation of highly reactive oxygen-containing molecules (i.e., reactive oxygen species [ROS]) that can damage other cell components; changes in the ratio of NADH to NAD+ (i.e., the cell’s redox state); tissue damage; fetal damage; impairment of other metabolic processes; cancer; and medication interactions. Several issues related to alcohol metabolism require further research. KEY WORDS: Ethanol-to acetaldehyde metabolism; alcohol dehydrogenase (ADH); aldehyde dehydrogenase (ALDH); acetaldehyde; acetate; cytochrome P450 2E1 (CYP2E1); catalase; reactive oxygen species (ROS); blood alcohol concentration (BAC); liver; stomach; brain; fetal alcohol effects; genetics and heredity; ethnic group; hypoxia The alcohol elimination rate varies state of liver cells. Chronic alcohol con- he effects of alcohol (i.e., ethanol) widely (i.e., three-fold) among individ- sumption and alcohol metabolism are on various tissues depend on its uals and is influenced by factors such as strongly linked to several pathological concentration in the blood T chronic alcohol consumption, diet, age, consequences and tissue damage. (blood alcohol concentration [BAC]) smoking, and time of day (Bennion and Understanding the balance of alcohol’s over time.