The Emerging Role for Cullin 4 Family of E3 Ligases in Tumorigenesis T ⁎ ⁎ Ji Chenga,B,1, Jianping Guob,1, Brian J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Escape from X Chromosome Inactivation Is an Intrinsic Property of the Jarid1c Locus
Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus Nan Lia,b and Laura Carrela,1 aDepartment of Biochemistry and Molecular Biology and bIntercollege Graduate Program in Genetics, Pennsylvania State College of Medicine, Hershey, PA 17033 Edited by Stanley M. Gartler, University of Washington, Seattle, WA, and approved September 23, 2008 (received for review August 8, 2008) Although most genes on one X chromosome in mammalian fe- Sequences on the X are hypothesized to propagate XCI (12) and males are silenced by X inactivation, some ‘‘escape’’ X inactivation to be depleted at escape genes (8, 9). LINE-1 repeats fit such and are expressed from both active and inactive Xs. How these predictions, particularly on the human X (8–10). Distinct dis- escape genes are transcribed from a largely inactivated chromo- tributions of other repeats classify some mouse X genes (5). some is not fully understood, but underlying genomic sequences X-linked transgenes also test the role of genomic sequences in are likely involved. We developed a transgene approach to ask escape gene expression. Most transgenes are X-inactivated, whether an escape locus is autonomous or is instead influenced by although a number escape XCI (e.g., refs. 13 and 14). Such X chromosome location. Two BACs carrying the mouse Jarid1c gene transgene studies indicate that, in addition to CTCF (6), locus and adjacent X-inactivated transcripts were randomly integrated control regions and matrix attachment sites are also not suffi- into mouse XX embryonic stem cells. Four lines with single-copy, cient to escape XCI (15, 16). -

DDB1 Targets Chk1 to the Cul4 E3 Ligase Complex in Normal Cycling Cells and in Cells Experiencing Replication Stress
Published OnlineFirst March 10, 2009; DOI: 10.1158/0008-5472.CAN-08-3382 Research Article DDB1 Targets Chk1 to the Cul4 E3 Ligase Complex in Normal Cycling Cells and in Cells Experiencing Replication Stress Van Leung-Pineda,1 Jiwon Huh,1 and Helen Piwnica-Worms1,2,3 Departments of 1Cell Biology and Physiology and 2Internal Medicine and 3Howard Hughes Medical Institute, Washington University School of Medicine, St. Louis, Missouri Abstract protein FANCE (8, 10–12). Chk1 carries out its functions both in the The Chk1 protein kinase preserves genome integrity in normal nucleus and at the centrosome (13). Drugs that block Chk1 kinase proliferating cells and in cells experiencing replicative and activity or enhance its proteolysis by interfering with binding to genotoxic stress. Chk1 is currently being targeted in antican- heat shock protein 90 (HSP90) are currently being tested as cer regimens. Here, we identify damaged DNA-binding protein anticancer agents (14–17). 1 (DDB1) as a novel Chk1-interacting protein. DDB1 is part of Chk1 is regulated by reversible phosphorylation and by an E3 ligase complex that includes the cullin proteins Cul4A ubiquitin-mediated proteolysis. Under periods of replicative stress, and Cul4B. We report that Cul4A/DDB1 negatively regulates the ATRIP/ATR module binds to single-stranded DNA and, Chk1 stability in vivo. Chk1 associates with Cul4A/DDB1 together with Rad17 and the 9-1-1 complex, activates Chk1 in a Claspin-dependent manner (18–22). ATR directly phosphorylates during an unperturbed cell division cycle and both Chk1 317 345 phosphorylation and replication stress enhanced these inter- Chk1 on two COOH-terminal residues: Ser (S317) and Ser actions. -

The Role and Mechanism of CRL4 E3 Ubiquitin Ligase in Cancer and Its Potential Therapy Implications
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No. 40 The role and mechanism of CRL4 E3 ubiquitin ligase in cancer and its potential therapy implications Youzhou Sang1,3,4, Fan Yan1,3,4 and Xiubao Ren2,3,4 1 Department of Immunology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, China 2 Department of Biotherapy, Tianjin Medical University Cancer Institute and Hospital, Tianjin, China 3 National Clinical Research Center of Cancer, Tianjin, China 4 Key Laboratory of Cancer Immunology and Biotherapy, Tianjin, China Correspondence to: Xiubao Ren, email: [email protected] Keywords: CRL4, CUL4, ubiquitination, cancer Received: April 08, 2015 Accepted: September 23, 2015 Published: October 09, 2015 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT CRLs (Cullin-RING E3 ubiquitin ligases) are the largest E3 ligase family in eukaryotes, which ubiquitinate a wide range of substrates involved in cell cycle regulation, signal transduction, transcriptional regulation, DNA damage response, genomic integrity, tumor suppression and embryonic development. CRL4 E3 ubiquitin ligase, as one member of CRLs family, consists of a RING finger domain protein, cullin4 (CUL4) scaffold protein and DDB1–CUL4 associated substrate receptors. The CUL4 subfamily includes two members, CUL4A and CUL4B, which share extensively sequence identity and functional redundancy. Aberrant expression of CUL4 has been found in a majority of tumors. Given the significance of CUL4 in cancer, understanding its detailed aspects of pathogenesis of human malignancy would have significant value for the treatment of cancer. -

Transcriptome Analyses of Rhesus Monkey Pre-Implantation Embryos Reveal A
Downloaded from genome.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press Transcriptome analyses of rhesus monkey pre-implantation embryos reveal a reduced capacity for DNA double strand break (DSB) repair in primate oocytes and early embryos Xinyi Wang 1,3,4,5*, Denghui Liu 2,4*, Dajian He 1,3,4,5, Shengbao Suo 2,4, Xian Xia 2,4, Xiechao He1,3,6, Jing-Dong J. Han2#, Ping Zheng1,3,6# Running title: reduced DNA DSB repair in monkey early embryos Affiliations: 1 State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China 2 Key Laboratory of Computational Biology, CAS Center for Excellence in Molecular Cell Science, Collaborative Innovation Center for Genetics and Developmental Biology, Chinese Academy of Sciences-Max Planck Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China 3 Yunnan Key Laboratory of Animal Reproduction, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, Yunnan 650223, China 4 University of Chinese Academy of Sciences, Beijing, China 5 Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan 650204, China 6 Primate Research Center, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, 650223, China * Xinyi Wang and Denghui Liu contributed equally to this work 1 Downloaded from genome.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press # Correspondence: Jing-Dong J. Han, Email: [email protected]; Ping Zheng, Email: [email protected] Key words: rhesus monkey, pre-implantation embryo, DNA damage 2 Downloaded from genome.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press ABSTRACT Pre-implantation embryogenesis encompasses several critical events including genome reprogramming, zygotic genome activation (ZGA) and cell fate commitment. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Genomic Approaches to Reproductive Disorders
Genomic Approaches to Reproductive Disorders Aleksandar Rajkovic Dept Obstetrics Gynecology and Reproductive Sciences University of Pittsburgh Magee Womens Research Institute Pittsburgh, PA Preconceptional Care Scope • Half of Pregnancies are Unintended • Medical Conditions • Mental Conditions • Immunization History • Nutritional Issues • Family History/Genetic Risk • Occupational/Environmental Exposures • Tobacco/Drug Abuse • Social Issues Preconceptional genetic screening Ethnic: Sickle cell disease Tay–Sachs disease Pan-ethnic: cystic fibrosis fragile X syndrome Spinal muscular atrophy Mendelian Inheritance • 5593 phenotypes for which molecular basis known • 3452 genes with phenotype causing mutation • Over 15,000 mutations to date known Preconceptional Pan Ethnic Testing • Screens for known mutations in more than 100 genes, easy on genetic counsellors • The screen is pan-ethnic • Useful also for couples undergoing IVF and potentially PGD • 1:5 will be carriers of a Mendelian disorder. • $600 (529 Euros) for the couple Genetic Counselling • Objective of the test • Test Methodology • Type of sample required (parents, siblings) • Possible outcomes (abnormal results, result of unknown clinical significance) ClinVar Stars and their interpretation Number of golden stars No submitter provided an interpretation with assertion criteria (no assertion criteria provided), none or no interpretation was provided (no assertion provided) At least one submitter provided an interpretation with assertion criteria (criteria provided, single submitter) -

Targeting the Methyltransferase SETD8 Impairs Tumor Cell Survival and Overcomes Drug Resistance Independently of P53 Status in Multiple Myeloma
bioRxiv preprint doi: https://doi.org/10.1101/776930; this version posted September 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Targeting the methyltransferase SETD8 impairs tumor cell survival and overcomes drug resistance independently of p53 status in multiple myeloma Laurie Herviou (1,2,4), Fanny Izard (3,4), Ouissem Karmous-Gadacha (2) , Claire Gourzones (1), Celine Bellanger (1), Eva Desmedt (5), Anqi Ma (6), Laure Vincent (7) , Guillaume Cartron (4,7), Karin Vanderkerken (5), Jian Jin (6), Elke De Bruyne (5), Charlotte Grimaud (3,4,8), Eric Julien (3,4,8 +) and Jérôme Moreaux (1,2,4+) (1) IGH, CNRS, Univ Montpellier, France (2) CHU Montpellier, Laboratory for Monitoring Innovative Therapies, Department of Biological Hematology, Montpellier, France (3) Institut de Recherche en Cancérologie de Montpellier (IRCM), INSERM U1194, Institut Régional du Cancer (ICM), Montpellier F-34298, France (4) University of Montpellier, Montpellier, F-34090, France (5) Department of Hematology and Immunology-Myeloma Center Brussels, Vrije Universiteit Brussel, Brussels, Belgium (6) Mount Sinai Center for Therapeutics Discovery, Departments of Pharmacological Sciences and Oncological Sciences, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, NeW York, NeW York 10029, United States. (7) CHU Montpellier, Department of Clinical Hematology, Montpellier, France (8) Centre National de la Recherche Scientifique (CNRS), F-34293, Montpellier, France + : co-last and corresponding authors ; corresponding authors: Eric Julien ([email protected]) and jérôme Moreaux ([email protected]). 1 bioRxiv preprint doi: https://doi.org/10.1101/776930; this version posted September 20, 2019. -

TRCP Promotes Cell Growth by Targeting PR-Set7/Set8 for Degradation
SCFβ-TRCP promotes cell growth by targeting PR-Set7/Set8 for degradation The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Wang, Z., X. Dai, J. Zhong, H. Inuzuka, L. Wan, X. Li, L. Wang, et al. 2015. “SCFβ-TRCP promotes cell growth by targeting PR-Set7/Set8 for degradation.” Nature Communications 6 (1): 10185. doi:10.1038/ ncomms10185. http://dx.doi.org/10.1038/ncomms10185. Published Version doi:10.1038/ncomms10185 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:23993465 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ARTICLE Received 4 May 2015 | Accepted 12 Nov 2015 | Published 15 Dec 2015 DOI: 10.1038/ncomms10185 OPEN SCFb-TRCP promotes cell growth by targeting PR-Set7/Set8 for degradation Zhiwei Wang1,2,*, Xiangpeng Dai2,*, Jiateng Zhong2,3,*, Hiroyuki Inuzuka2, Lixin Wan2, Xiaoning Li2,4, Lixia Wang1, Xiantao Ye1, Liankun Sun4, Daming Gao2,5,LeeZou6 & Wenyi Wei2 The Set8/PR-Set7/KMT5a methyltransferase plays critical roles in governing transcriptional regulation, cell cycle progression and tumorigenesis. Although CRL4Cdt2 was reported to regulate Set8 stability, deleting the PIP motif only led to partial resistance to ultraviolet- induced degradation of Set8, indicating the existence of additional E3 ligase(s) controlling Set8 stability. Furthermore, it remains largely undefined how DNA damage-induced kinase cascades trigger the timely destruction of Set8 to govern tumorigenesis. -

Hyperphosphorylation Repurposes the CRL4B E3 Ligase to Coordinate Mitotic Entry and Exit
Research Collection Doctoral Thesis Identification and Characterization of Novel Cullin 4-based E3 Ligases in Somatic Cell Division Author(s): da Graça Gilberto, Samuel Filipe Publication Date: 2017 Permanent Link: https://doi.org/10.3929/ethz-b-000219014 Rights / License: In Copyright - Non-Commercial Use Permitted This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library DISS. ETH NO. 24583 Identification and characterization of novel Cullin 4-based E3 ligases in somatic cell division A thesis submitted to attain the degree of DOCTOR OF SCIENCES of ETH ZURICH (Dr. sc. ETH Zurich) Presented by SAMUEL FILIPE DA GRAÇA GILBERTO MSc in Biochemistry, University of Lisbon Born on 21.02.1988 Citizen of Portugal Accepted on the recommendation of Prof. Dr. Matthias Peter Prof. Dr. Anton Wutz 2017 Table of contents 1. General introduction ..................................................................................................................1 1.1. The ubiquitylation machinery ....................................................................................................... 1 1.2. Principles of cell cycle regulation: a focus on CRLs and the APC/C .............................................. 3 1.3. Cullin-4 RING E3 ligases: cell-cycle regulation and beyond ........................................................ 14 1.4. Functional distinctions between CUL4A and CUL4B .................................................................. -

And CAND1-Dependent Remodelling of the Budding Yeast SCF Complex
ARTICLE Received 31 Jan 2013 | Accepted 20 Feb 2013 | Published 27 Mar 2013 DOI: 10.1038/ncomms2628 OPEN CSN- and CAND1-dependent remodelling of the budding yeast SCF complex Aleksandra Zemla1, Yann Thomas1, Sylwia Kedziora1, Axel Knebel1, Nicola T Wood1, Gwenae¨l Rabut2 & Thimo Kurz1 Cullin–RING ligases (CRLs) are ubiquitin E3 enzymes with variable substrate-adaptor and -receptor subunits. All CRLs are activated by modification of the cullin subunit with the ubiquitin-like protein Nedd8 (neddylation). The protein CAND1 (Cullin-associated-Nedd8- dissociated-1) also promotes CRL activity, even though it only interacts with inactive ligase complexes. The molecular mechanism underlying this behaviour remains largely unclear. Here, we find that yeast SCF (Skp1–Cdc53–F-box) Cullin–RING complexes are remodelled in a CAND1-dependent manner, when cells are switched from growth in fermentable to non-fermentable carbon sources. Mechanistically, CAND1 promotes substrate adaptor release following SCF deneddylation by the COP9 signalosome (CSN). CSN- or CAND1- mutant cells fail to release substrate adaptors. This delays the formation of new complexes during SCF reactivation and results in substrate degradation defects. Our results shed light on how CAND1 regulates CRL activity and demonstrate that the cullin neddylation– deneddylation cycle is not only required to activate CRLs, but also to regulate substrate specificity through dynamic substrate adaptor exchange. 1 Scottish Institute for Cell Signalling, Protein Ubiquitylation Unit, College of Life Sciences, University of Dundee, Dow Street, Dundee DD1 5EH, Scotland, UK. 2 CNRS, Universite´ Rennes 1, Institut de Ge´ne´tique et De´veloppement de Rennes, 2 avenue du Professeur Le´on Bernard, CS 34317, Rennes Cedex 35043, France. -
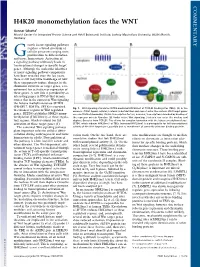
H4K20 Monomethylation Faces the WNT
COMMENTARY H4K20 monomethylation faces the WNT Gunnar Schotta1 Munich Center for Integrated Protein Science and Adolf Butenandt Institute, Ludwig Maximilians University, 80336 Munich, Germany rowth factor signaling pathways regulate a broad spectrum of G cellular processes ranging from proliferation to differentiation and tissue homeostasis. Activation of a signaling pathway ultimately leads to transcriptional changes in specific target genes. Although the molecular identities of many signaling pathway components have been revealed over the last years, there is still very little knowledge of how these components induce changes in the chromatin structure of target genes, a re- quirement for activation or repression of these genes. A new link is provided by an interesting paper in PNAS that demon- strates that in the context of Wnt signaling the histone methyltransferase SETD8 (PR-SET7, KMT5a, SET8) is recruited Fig. 1. Wnt signaling stimulates SETD8-mediated H4K20me1 at TCF/LEF binding sites (TBEs). (A)Inthe to enhancer regions of Wnt-regulated absence of Wnt ligand, cellular β-catenin is destabilized and cannot enter the nucleus. Wnt target genes genes. SETD8 establishes H4K20 mono- are constitutively bound by TCF/LEF transcription factors; however, transcription is blocked by binding of methylation (H4K20me1) at these regula- the repressor protein Groucho. (B) Under active Wnt signaling, β-catenin can enter the nucleus and tory regions, which is crucial for full displace Groucho from TCF/LEF. This allows for complex formation with the histone methyltransferase activation of these target genes (1). SETD8, which induces H4K20me1 at TBEs. Increased H4K20me1 is a prerequisite for full transcriptional The canonical Wnt signaling pathway activity of the Wnt target gene, possibly due to recruitment of currently unknown binding proteins. -

Deletion of DDB1- and CUL4- Associated Factor-17 (Dcaf17) Gene
www.nature.com/scientificreports OPEN Deletion of DDB1- and CUL4- associated factor-17 (Dcaf17) gene causes spermatogenesis defects Received: 27 October 2017 Accepted: 31 May 2018 and male infertility in mice Published: xx xx xxxx Asmaa Ali1, Bhavesh V. Mistry1, Hala A. Ahmed1, Razan Abdulla1, Hassan A. Amer3, Abdelbary Prince3, Anas M. Alazami2, Fowzan S. Alkuraya 2 & Abdullah Assiri 1,4,5 DDB1– and CUL4–associated factor 17 (Dcaf17) is a member of DCAF family genes that encode substrate receptor proteins for Cullin-RING E3 ubiquitin ligases, which play critical roles in many cellular processes. To unravel the function of DCAF17, we performed expression profling of Dcaf17 in diferent tissues of wild type mouse by qRT-PCR and generated Dcaf17 knockout mice by gene targeting. Expression profling of Dcaf17 showed highest expression in testis. Analyses of Dcaf17 transcripts during post-natal development of testis at diferent ages displayed gradual increase in Dcaf17 mRNA levels with the age. Although Dcaf17 disruption did not have any efect on female fertility, Dcaf17 deletion led to male infertility due to abnormal sperm development. The Dcaf17−/− mice produced low number of sperm with abnormal shape and signifcantly low motility. Histological examination of the Dcaf17−/− testis revealed impaired spermatogenesis with presence of vacuoles and sloughed cells in the seminiferous tubules. Disruption of Dcaf17 caused asymmetric acrosome capping, impaired nuclear compaction and abnormal round spermatid to elongated spermatid transition. For the frst time, these data indicate that DCAF17 is essential for spermiogenesis. Infertility is a global health problem that afects 15% of couples worldwide1,2. Almost half of these infertility cases are attributed to male infertility, where about 75% of all the male factor infertility attributed to defective sper- matogenesis3–5.