Population Genetic Structure of Iris Pumila L. in Ukraine: Effects of Habitat Fragmentation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Qrno. 1 2 3 4 5 6 7 1 CP 2903 77 100 0 Cfcl3
QRNo. General description of Type of Tariff line code(s) affected, based on Detailed Product Description WTO Justification (e.g. National legal basis and entry into Administration, modification of previously the restriction restriction HS(2012) Article XX(g) of the GATT, etc.) force (i.e. Law, regulation or notified measures, and other comments (Symbol in and Grounds for Restriction, administrative decision) Annex 2 of e.g., Other International the Decision) Commitments (e.g. Montreal Protocol, CITES, etc) 12 3 4 5 6 7 1 Prohibition to CP 2903 77 100 0 CFCl3 (CFC-11) Trichlorofluoromethane Article XX(h) GATT Board of Eurasian Economic Import/export of these ozone destroying import/export ozone CP-X Commission substances from/to the customs territory of the destroying substances 2903 77 200 0 CF2Cl2 (CFC-12) Dichlorodifluoromethane Article 46 of the EAEU Treaty DECISION on August 16, 2012 N Eurasian Economic Union is permitted only in (excluding goods in dated 29 may 2014 and paragraphs 134 the following cases: transit) (all EAEU 2903 77 300 0 C2F3Cl3 (CFC-113) 1,1,2- 4 and 37 of the Protocol on non- On legal acts in the field of non- _to be used solely as a raw material for the countries) Trichlorotrifluoroethane tariff regulation measures against tariff regulation (as last amended at 2 production of other chemicals; third countries Annex No. 7 to the June 2016) EAEU of 29 May 2014 Annex 1 to the Decision N 134 dated 16 August 2012 Unit list of goods subject to prohibitions or restrictions on import or export by countries- members of the -
(A) Journals with the Largest Number of Papers Reporting Estimates Of
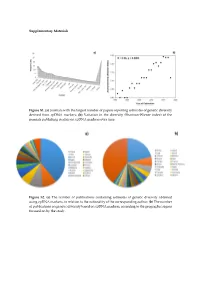
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

José Guadalupe García-Franco
CURRICULUM VITAE JOSÉ GUADALUPE GARCÍA-FRANCO 20/08/2015 Curriculun Vitae García-Franco Contenido 1. DATOS PERSONALES ..................................................................................................................... 5 2. DATOS LABORALES ....................................................................................................................... 5 3. FORMACIÓN PROFESIONAL ............................................................................................................ 5 3.1. Licenciatura: .......................................................................................................................... 5 3.2. Maestría: ................................................................................................................................ 5 3.3. Doctorado: ............................................................................................................................. 5 4. PERTENENCIA AL SISTEMA NACIONAL DE INVESTIGADORES ......................................................... 5 5. DOMINIO DE IDIOMAS EXTRANJEROS ............................................................................................ 5 6. BECAS OBTENIDAS PARA SU FORMACIÓN PROFESIONAL ............................................................... 5 7. EXPERIENCIA LABORAL ................................................................................................................ 6 8. ASISTENCIA A CURSOS Y TALLERES DE CAPACITACIÓN .............................................................. 6 9. -

Vol. 49 Valencia, X-2011 FLORA MONTIBERICA
FLORA MONTIBERICA Publicación periódica especializada en trabajos sobre la flora del Sistema Ibérico Vol. 49 Valencia, X-2011 FLORA MONTIBERICA Publicación independiente sobre temas relacionados con la flora y la vegetación (plantas vasculares) de la Península Ibérica, especialmente de la Cordillera Ibérica y tierras vecinas. Fundada en diciembre de 1995, se publican tres volúmenes al año con una periodicidad cuatrimestral. Editor y Redactor general: Gonzalo Mateo Sanz. Jardín Botánico. Universidad de Valencia. C/ Quart, 80. E-46008 Valencia. Redactores adjuntos: Javier Fabado Alós. Redactor página web y editor adjunto: José Luis Benito Alonso. Edición en Internet: www.floramontiberica.org Flora Montiberica.org es la primera revista de botánica en español que ofrece de forma gratuita todos sus contenidos a través de la red. Consejo editorial: Antoni Aguilella Palasí (Universidad de Valencia) Juan A. Alejandre Sáenz (Herbarium Alejandre, Vitoria) Vicente J. Arán Redó (Consejo Superior de Investigaciones Científicas, Madrid) Manuel Benito Crespo Villalba (Universidad de Alicante) José María de Jaime Lorén (Universidad Cardenal Herrera-CEU, Moncada) Emilio Laguna Lumbreras ((Departamento de Medio Ambiente. Gobierno de la Comunidad Valenciana) Pedro Montserrat Recoder (Consejo Superior de Investigaciones Científicas, Jaca). Edita: Flora Montiberica. Valencia (España). ISSN: 1138-5952 – ISSN edición internet: 1988-799X. Depósito Legal: V-5097-1995. Portada: Ophioglossum azoricum C. Presl, procedente de Sotorribas (Cuenca). Véase pág. 36 de este número. Flora Montiberica 49: 3-5 (X-2011). ISSN 1988-799X NUEVA LOCALIDAD VALENCIANA DE PUCCINELLIA HISPANICA JULIÀ & J. M. MONTSERRAT (POACEAE) P. Pablo FERRER GALLEGO1 & Roberto ROSELLÓ GIMENO2 1Servicio de Biodiversidad, Centro para la Investigación y la Experimentación Forestal de la Generalitat Valenciana (CIEF). -

CBD First National Report
FIRST NATIONAL REPORT OF THE REPUBLIC OF SERBIA TO THE UNITED NATIONS CONVENTION ON BIOLOGICAL DIVERSITY July 2010 ACRONYMS AND ABBREVIATIONS .................................................................................... 3 1. EXECUTIVE SUMMARY ........................................................................................... 4 2. INTRODUCTION ....................................................................................................... 5 2.1 Geographic Profile .......................................................................................... 5 2.2 Climate Profile ...................................................................................................... 5 2.3 Population Profile ................................................................................................. 7 2.4 Economic Profile .................................................................................................. 7 3 THE BIODIVERSITY OF SERBIA .............................................................................. 8 3.1 Overview......................................................................................................... 8 3.2 Ecosystem and Habitat Diversity .................................................................... 8 3.3 Species Diversity ............................................................................................ 9 3.4 Genetic Diversity ............................................................................................. 9 3.5 Protected Areas .............................................................................................10 -

12.2% 122,000 135M Top 1% 154 4,800
View metadata, citation and similar papers at core.ac.uk brought to you by CORE We are IntechOpen, provided by IntechOpen the world’s leading publisher of Open Access books Built by scientists, for scientists 4,800 122,000 135M Open access books available International authors and editors Downloads Our authors are among the 154 TOP 1% 12.2% Countries delivered to most cited scientists Contributors from top 500 universities Selection of our books indexed in the Book Citation Index in Web of Science™ Core Collection (BKCI) Interested in publishing with us? Contact [email protected] Numbers displayed above are based on latest data collected. For more information visit www.intechopen.com 18 Living on the Edge: Various Modes of Persistence at the Range Margins of Some Far Eastern Species Elena Artyukova, Marina Kozyrenko, Olga Koren, Alla Kholina, Olga Nakonechnaya and Yuri Zhuravlev Institute of Biology and Soil Science, Far East Branch of Russian Academy of Sciences Russia 1. Introduction Present-day patterns of plant distribution have been formed under the influence of various biotic and abiotic factors. Plant distribution reflects the habitat preferences of species and the outcome of their competition as well as the complex evolutionary processes resulting in the specificity of mating systems, the genetic structure of different species and other aspects of species biology. Together, these factors determine the current ranges and distributions of plant species. At the edge of a species' range, the significance of particular interactions with the environment becomes more pronounced. However, our understanding of this class of interactions is limited. There is debate about whether these interactions represent a distinct and ordered set of related phenomena or whether they are unrelated and without order. -

Various Modes of Persistence at the Range Margins of Some Far Eastern Species
18 Living on the Edge: Various Modes of Persistence at the Range Margins of Some Far Eastern Species Elena Artyukova, Marina Kozyrenko, Olga Koren, Alla Kholina, Olga Nakonechnaya and Yuri Zhuravlev Institute of Biology and Soil Science, Far East Branch of Russian Academy of Sciences Russia 1. Introduction Present-day patterns of plant distribution have been formed under the influence of various biotic and abiotic factors. Plant distribution reflects the habitat preferences of species and the outcome of their competition as well as the complex evolutionary processes resulting in the specificity of mating systems, the genetic structure of different species and other aspects of species biology. Together, these factors determine the current ranges and distributions of plant species. At the edge of a species' range, the significance of particular interactions with the environment becomes more pronounced. However, our understanding of this class of interactions is limited. There is debate about whether these interactions represent a distinct and ordered set of related phenomena or whether they are unrelated and without order. Different approaches to this problem are needed in different situations. Understanding the processes of microevolution in species at the edges of their ranges is of great interest, particularly in view of the continuing decline in worldwide biodiversity and ongoing and future climate changes. When the area of a plant's habitat is sufficient, most populations exist in a relatively stable condition, and changes in their genetic structure follow slow processes, such as gene flow and genetic drift. However, in populations growing at the edge of their range, the rates of genetic processes can change dramatically. -

Protection of the Taiga–Steppe Ecosystems of the Western Shore of Lake Baikal
A.P. Sizykh, Int. J. of Design & Nature and Ecodynamics. Vol. 4, No. 1 (2009) 66–71 SHORT COMMUNICATION PROTECTION OF THE TAIGA–STEPPE ECOSYSTEMS OF THE WESTERN SHORE OF LAKE BAIKAL A.P. SIZYKH Siberian Institute of Plant Physiology and Biochemistry, Russian Academy of Sciences, Irkutsk, Russia. ABSTRACT By monitoring the spatial–coenotic changes of taiga–steppe communities and fl uctuations of climatic factors over the course of many years, it was possible to identify dynamic trends of the region’s taiga–steppe ecosys- tems’ formation. The layered character is most clearly pronounced, where meadow–steppe and forest–steppe plant species become predominant, irrespective of the type of habitat of the communities. The composition of taiga–steppe (transitional between forest and steppe) ecosystems reveals a spatial spreading of sinuosities of mosses characteristic for the light-coniferous and for the dark-coniferous taiga. A special trait of the spatial changes in the region’s vegetation structure should be mentioned, namely, a tendency toward a very active intrusion of wood species: pine and larch, into steppe communities thus forming curtains of uneven-age trees ranging from shoots to 25- to 30-year trees. Isolated (amidst steppes) trees as old as 40–50 years were reported previously. Processes of overall mesophytization and aforestation of steppes were observed throughout the western shore are of Lake Baikal. The genesis of the region’s vegetation over the course of Holocene and mod- ern tendencies of the communities at the background of the changes of temperature–hydrological regimes over the last several decades suggest the initial stage of formation here of taiga–steppe ecosystems characteristic only for this region of east Siberia. -

Plastid DNA Variation of the Endemic Species Oxytropis Glandulosa Turcz
Turkish Journal of Botany Turk J Bot (2018) 42: 38-50 http://journals.tubitak.gov.tr/botany/ © TÜBİTAK Research Article doi:10.3906/bot-1706-11 Plastid DNA variation of the endemic species Oxytropis glandulosa Turcz. (Fabaceae) 1, 1 1 2 3 2 Alla KHOLINA *, Marina KOZYRENKO , Elena ARTYUKOVA , Denis SANDANOV , Inessa SELYUTINA , Daba CHIMITOV 1 Federal Scientific Center of the East Asia Terrestrial Biodiversity, Far Eastern Branch of the Russian Academy of Sciences, Vladivostok, Russia 2 Institute of General and Experimental Biology, Siberian Branch of the Russian Academy of Sciences, Ulan-Ude, Russia 3 Central Siberian Botanical Garden, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia Received: 06.06.2017 Accepted/Published Online: 15.09.2017 Final Version: 11.01.2018 Abstract: Oxytropis glandulosa Turcz. (Fabaceae) is a rare perennial plant endemic to Buryatia (Russia). We sequenced three intergenic spacers (the psbA–trnH, trnL–trnF, and trnS–trnG) of chloroplast DNA (cpDNA) in specimens of four populations from two geographic regions (Barguzin and Yeravna depressions) within this species range. The levels of haplotype and nucleotide diversity varied in the range of 0.133–0.911 and 0.0002–0.0059, respectively. The highest values of these parameters are characteristic of populations located in the Barguzin depression. Variable sites detected within the intergenic spacers allowed the identification of eleven haplotypes; nine of them were found in the populations from the Barguzin depression and only two haplotypes were found in the populations from the Yeravna depression. No common haplotypes for all four populations were found. A high level of population differentiation (ΦST = 0.758, P < 0.0001) and the absence of evidence for isolation by distance were also found in O. -

Особенности Морфологии Семян Некоторых Видов Рода Iris (Iridaceae) Юга Западной Сибири Е.Н
НАУЧНЫЙ ЖУРНАЛ РАСТИТЕЛЬНЫЙ МИР АЗИАТСКОЙ РОССИИ Растительный мир Азиатской России, 2012, № 1(9), с. 44–49 h p://www.izdatgeo.ru УДК 582.572.7:581.48(571.1/.5) ОСОБЕННОСТИ МОРФОЛОГИИ СЕМЯН НЕКОТОРЫХ ВИДОВ РОДА IRIS (IRIDACEAE) ЮГА ЗАПАДНОЙ СИБИРИ Е.Н. Кайгородова, О.В. Дорогина, Т.В. Елисафенко Центральный сибирский ботанический сад СО РАН, 630090, Новосибирск, ул. Золотодолинская, 101, e-mail: [email protected]; [email protected]; [email protected] Исследованы морфологические признаки и структура поверхности семян рода Iris L. с помощью сканирую- щего электронного микроскопа. У семян I. humilis Georgi выражен присемянник, у I. tigridia Bunge и I. pota- ninii Maxim. присемянник незначителен, а у семян I. glaucescens Bunge он не обнаружен. Обработка паром семян I. glaucescens, характеризующихся плотной кутикулой, привела к ее элиминации. Ключевые слова: Iridaceae, Iris, структура поверхности семян, морфологические признаки. MORPHOLOGY FEATURES OF SEEDS IN SOME SPECIES IRIS (IRIDACEAE) OF THE SOUTH OF WESTERN SIBERIA E.N. Kaygorodova, O.V. Dorogina, T.V. Elisafenko Central Siberian Botanical Garden, SB RAS, 630090, Novosibirsk, Zolotodolinskaya str., 101, e-mail: [email protected]; [email protected]; [email protected] Morphology features and seed surface of coat structure of the genus Iris L. were examined using the scaning electron microscope. In seeds of I. humilis present well expressed aril, in seeds of I. tigridia and I. potaninii it is poorly formed but for seeds of I. glaucescens it was absent. Treatment of steam I. glaucescens seeds having of thik cuticle led to the elimination of cuticle. Key words: Iridaceae, Iris, seed surface of coat structure, morphology features. -

Natura 2000 Interpretation Manual of European Union
NATURA 2000 INTERPRETATION MANUAL OF EUROPEAN UNION HABITATS Version EUR 15 Q) .c Ol c: 0 "iii 0 ·"'a <>c: ~ u.. C: ~"' @ *** EUROPEAN COMMISSION ** ** DGXI ... * * Environment, Nuclear Security and Civil Protection 0 *** < < J J ) NATURA 2000 INTERPRETATION MANUAL OF EUROPEAN UNION HABITATS Version EUR 15 This 111anual is a scientific reference document adopted by the habitats committee on 25 April 1996 Compiled by : Carlos Romio (DG. XI • 0.2) This document is edited by Directorate General XI "Environment, Nuclear Safety and Civil Protection" of the European Commission; author service: Unit XI.D.2 "Nature Protection, Coastal Zones and Tourism". 200 rue de Ia Loi, B-1049 Bruxelles, with the assistance of Ecosphere- 3, bis rue des Remises, F-94100 Saint-Maur-des-Fosses. Neither the European Commission, nor any person acting on its behalf, is responsible for the use which may be made of this document. Contents WHY THIS MANUAL?---------------- 1 Historical review ............................................... 1 The Manual .................................................... 1 THE EUR15 VERSION 3 Biogeographical regions .......................................... 3 Vegetation levels ................................................ 4 Explanatory notes ............................................... 5 COASTAL AND HALOPHYTIC HABITATS 6 Open sea and tidal areas . 6 Sea cliffs and shingle or stony beaches ............................ 10 Atlantic and continental salt marshes and salt meadows . 12 Mediterranean and thermo-Atlantic saltmarshes and salt meadows .... 14 Salt and gypsum continental steppes . 15 COASTAL SAND DUNES AND CONTINENTAL DUNES 17 Sea dunes of the Atlantic, North Sea and Baltic coasts ............... 17 Sea dunes of the Mediterranean coast . 22 Continental dunes, old and decalcified . 24 FRESHWATER HABITATS 26 Standing water . 26 Running water . 29 TEMPERATE HEATH AND SCRUB------------ 33 SCLEROPHYLLOUS SCRUB (MATORRAL) 40 Sub-Mediterranean and temperate . -

Phytochemical Analysis and Total Antioxidant Capacity of Rhizome, Above-Ground Vegetative Parts and Flower of Three Iris Species
Title: Phytochemical analysis and total antioxidant capacity of rhizome, above-ground vegetative parts and flower of three Iris species Authors: Aleksandar Ž. Kostić, Uroš M. Gašić, Mirjana B. Pešić, Sladjana P. Stanojević, Miroljub B. Barać, Marina P. Mačukanović-Jocić, Stevan N. Avramov, and Živoslav Lj. Tešić This manuscript has been accepted after peer review and appears as an Accepted Article online prior to editing, proofing, and formal publication of the final Version of Record (VoR). This work is currently citable by using the Digital Object Identifier (DOI) given below. The VoR will be published online in Early View as soon as possible and may be different to this Accepted Article as a result of editing. Readers should obtain the VoR from the journal website shown below when it is published to ensure accuracy of information. The authors are responsible for the content of this Accepted Article. To be cited as: Chem. Biodiversity 10.1002/cbdv.201800565 Link to VoR: http://dx.doi.org/10.1002/cbdv.201800565 Chemistry & Biodiversity 10.1002/cbdv.201800565 Chem. Biodiversity Phytochemical analysis and total antioxidant capacity of rhizome, above-ground vegetative parts and flower of three Iris species Aleksandar Ž. Kostića*, Uroš M. Gašićb, Mirjana B. Pešića, Sladjana P. Stanojevića, Miroljub B. Baraća, Marina P. Mačukanović-Jocićc, Stevan N. Avramovd, Živoslav Lj. Tešićb a University of Belgrade, Faculty of Agriculture, Chair of Chemistry and Biochemistry, Nemanjina 6, 11080, Belgrade, Serbia *Corresponding author E-mail address: [email protected]