Author Index
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Financial Supporters
FINANCIAL SUPPORTERS Care and Share Food Bank for Southern Colorado 2011-12 Financial Supporters 100% Chiropractic Lanny and Paul Adams Mr. and Mrs. Jeffrey Ahrendsen 14K Real Estate Investments LLC Ms. Laura Adams Mr. Kevin Ahrens 1882 Management Mr. and Mrs. Lon Adams Mr. and Mrs. Charles Aiken 1st Cavalry Rocky Mountain Chapter Col and Mrs. Louis Adams Ms. Laverne Ainley 221 South Oak Bistro Ms. Maggie Adams Air Academy Federal Credit Union 4-Bits 4-H Club Ms. Mary Adams Air Academy Federal Credit Union 4Clicks - Solutions, LLC Mr. Michael Adams Air Academy High School - District 20 A & L Aluminum Manufacturing Company Mr. and Mrs. Rexford Adams Mr. TJ Airhart A Handymike Home Repair Mr. and Mrs. Robert Adams Aka Wilson, LLC A to Z Realty Mr. S. Michael Adams Mr. Richard Alaniz AA “Accurate and Affordable” Striping, Inc. Mr. and Mrs. Samuel Adams Ms. Susan Alarid AAA NCNU Insurance Exchange Mr. Steve Adams Ms. Karin Alaska AAA Northern California Nevada & Utah Suzanne Adams Mr. Arturo Albanesi AARP Foundation Adams Bank & Trust Mr. and Mrs. Mac Alberico Ms. Renee Abbe Mrs. Alda Adcox Ms. Cheryl Alberto Ms. Marjory Abbott Add Staff Inc. Mr. and Mrs. Dewey Albertson Jr. Mr. and Mrs. Peyton Abbott Ms. Constance Addington Mr. W. Gary Albertson Ms. Stephanie Abbott Mr. and Mrs. D. V. Addington Albertsons LLC Ms. Brianna Abby Ms. Linda Addington Mr. and Mrs. Albert Albrandt Mr. and Mrs. Donald Abdallah Ms. Vicky Addison Mr. Gerald Albrecht Mr. Tony Abdella Ms. Deirdre Aden-Smith Ms. Patricia Albright Mr. and Mrs. William Abel Mr. -
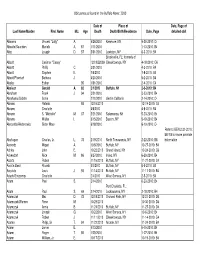
Obituaries Buffalo News 2010 by Name
Obituaries as found in the Buffalo News: 2010 Date of Place of Date, Page of Last Name/Maiden First Name M.I. Age Death Death/Birth/Residence Date, Page detailed obit Abbarno Vincent "Lolly" A. 9/26/2010 Kenmore, NY 9-30-2010: C4 Abbatte/Saunders Murielle A. 87 1/11/2010 1-13-2010: B4 Abbo Joseph D. 57 5/31/2010 Lewiston, NY 6-3-2010: B4 Brooksville, FL; formerly of Abbott Casimer "Casey" 12/19/22009 Cheektowaga, NY 4-18-2010: C6 Abbott Phillip C. 3/31/2010 4-3-2010: B4 Abbott Stephen E. 7/6/2010 7-8-2010: B4 Abbott/Pfoetsch Barbara J. 4/20/2010 5-2-2010: B4 Abeles Esther 95 1/31/2010 2-4-2010: C4 Abelson Gerald A. 82 2/1/2010 Buffalo, NY 2-3-2010: B4 Abraham Frank J. 94 3/21/2010 3-23-2010: B4 Abrahams/Gichtin Sonia 2/10/2010 died in California 2-14-2010: C4 Abramo Rafeala 93 12/16/2010 12-19-2010: C4 Abrams Charlotte 4/6/2010 4-8-2010: B4 Abrams S. "Michelle" M. 37 5/21/2010 Salamanca, NY 5-23-2010: B4 Abrams Walter I. 5/15/2010 Basom, NY 5-19-2010: B4 Abrosette/Aksterowicz Sister Mary 6/18/2010 6-19-2010: C4 Refer to BEN 2-21-2010: B6/7/8 for more possible Abshagen Charles, Jr. L. 73 2/19/2010 North Tonawanda, NY 2-22-2010: B8 information Acevedo Miguel A. 10/6/2010 Buffalo, NY 10-27-2010: B4 Achkar John E. -

(“Spider-Man”) Cr
PRIVILEGED ATTORNEY-CLIENT COMMUNICATION EXECUTIVE SUMMARY SECOND AMENDED AND RESTATED LICENSE AGREEMENT (“SPIDER-MAN”) CREATIVE ISSUES This memo summarizes certain terms of the Second Amended and Restated License Agreement (“Spider-Man”) between SPE and Marvel, effective September 15, 2011 (the “Agreement”). 1. CHARACTERS AND OTHER CREATIVE ELEMENTS: a. Exclusive to SPE: . The “Spider-Man” character, “Peter Parker” and essentially all existing and future alternate versions, iterations, and alter egos of the “Spider- Man” character. All fictional characters, places structures, businesses, groups, or other entities or elements (collectively, “Creative Elements”) that are listed on the attached Schedule 6. All existing (as of 9/15/11) characters and other Creative Elements that are “Primarily Associated With” Spider-Man but were “Inadvertently Omitted” from Schedule 6. The Agreement contains detailed definitions of these terms, but they basically conform to common-sense meanings. If SPE and Marvel cannot agree as to whether a character or other creative element is Primarily Associated With Spider-Man and/or were Inadvertently Omitted, the matter will be determined by expedited arbitration. All newly created (after 9/15/11) characters and other Creative Elements that first appear in a work that is titled or branded with “Spider-Man” or in which “Spider-Man” is the main protagonist (but not including any team- up work featuring both Spider-Man and another major Marvel character that isn’t part of the Spider-Man Property). The origin story, secret identities, alter egos, powers, costumes, equipment, and other elements of, or associated with, Spider-Man and the other Creative Elements covered above. The story lines of individual Marvel comic books and other works in which Spider-Man or other characters granted to SPE appear, subject to Marvel confirming ownership. -
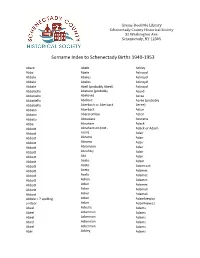
Surname Index to Schenectady Births 1940-1953
Grems-Doolittle Library Schenectady County Historical Society 32 Washington Ave. Schenectady, NY 12305 Surname Index to Schenectady Births 1940-1953 Abare Abele Ackley Abba Abele Ackroyd Abbale Abeles Ackroyd Abbale Abeles Ackroyd Abbale Abell (probably Abeel) Ackroyd Abbatiello Abelone (probably Acord Abbatiello Abelove) Acree Abbatiello Abelove Acree (probably Abbatiello Aberbach or Aberback Aeree) Abbato Aberback Acton Abbato Abercrombie Acton Abbato Aboudara Acucena Abbe Abraham Adack Abbott Abrahamson (not - Adack or Adach Abbott nson) Adair Abbott Abrams Adair Abbott Abrams Adair Abbott Abramson Adair Abbott Abrofsky Adair Abbott Abt Adair Abbott Aceto Adam Abbott Aceto Adamczak Abbott Aceto Adamec Abbott Aceto Adamec Abbott Acken Adamec Abbott Acker Adamec Abbott Acker Adamek Abbott Acker Adamek Abbzle = ? spelling Acker Adamkiewicz unclear Acker Adamkiewicz Abeel Ackerle Adams Abeel Ackerman Adams Abeel Ackerman Adams Abeel Ackerman Adams Abeel Ackerman Adams Abel Ackley Adams Grems-Doolittle Library Schenectady County Historical Society 32 Washington Ave. Schenectady, NY 12305 Surname Index to Schenectady Births 1940-1953 Adams Adamson Ahl Adams Adanti Ahles Adams Addis Ahman Adams Ademec or Adamec Ahnert Adams Adinolfi Ahren Adams Adinolfi Ahren Adams Adinolfi Ahrendtsen Adams Adinolfi Ahrendtsen Adams Adkins Ahrens Adams Adkins Ahrens Adams Adriance Ahrens Adams Adsit Aiken Adams Aeree Aiken Adams Aernecke Ailes = ? Adams Agans Ainsworth Adams Agans Aker (or Aeher = ?) Adams Aganz (Agans ?) Akers Adams Agare or Abare = ? Akerson Adams Agat Akin Adams Agat Akins Adams Agen Akins Adams Aggen Akland Adams Aggen Albanese Adams Aggen Alberding Adams Aggen Albert Adams Agnew Albert Adams Agnew Albert or Alberti Adams Agnew Alberti Adams Agostara Alberti Adams Agostara (not Agostra) Alberts Adamski Agree Albig Adamski Ahave ? = totally Albig Adamson unclear Albohm Adamson Ahern Albohm Adamson Ahl Albohm (not Albolm) Adamson Ahl Albrezzi Grems-Doolittle Library Schenectady County Historical Society 32 Washington Ave. -

Sep CUSTOMER ORDER FORM
OrdErS PREVIEWS world.com duE th 18 FeB 2013 Sep COMIC THE SHOP’S pISpISRReeVV eeWW CATALOG CUSTOMER ORDER FORM CUSTOMER 601 7 Sep13 Cover ROF and COF.indd 1 8/8/2013 9:49:07 AM Available only DOCTOR WHO: “THE NAME OF from your local THE DOCTOR” BLACK T-SHIRT comic shop! Preorder now! GODZILLA: “KING OF THANOS: THE SIMPSONS: THE MONSTERS” “INFINITY INDEED” “COMIC BOOK GUY GREEN T-SHIRT BLACK T-SHIRT COVER” BLUE T-SHIRT Preorder now! Preorder now! Preorder now! 9 Sep 13 COF Apparel Shirt Ad.indd 1 8/8/2013 9:54:50 AM Sledgehammer ’44: blaCk SCienCe #1 lightning War #1 image ComiCS dark horSe ComiCS harleY QUinn #0 dC ComiCS Star WarS: Umbral #1 daWn of the Jedi— image ComiCS forCe War #1 dark horSe ComiCS Wraith: WelCome to ChriStmaSland #1 idW PUbliShing BATMAN #25 CATAClYSm: the dC ComiCS ULTIMATES’ laSt STAND #1 marvel ComiCS Sept13 Gem Page ROF COF.indd 1 8/8/2013 2:53:56 PM Featured Items COMIC BOOKS & GRAPHIC NOVELS Shifter Volume 1 HC l AnomAly Productions llc The Joyners In 3D HC l ArchAiA EntErtAinmEnt llc All-New Soulfire #1 l AsPEn mlt inc Caligula Volume 2 TP l AvAtAr PrEss inc Shahrazad #1 l Big dog ink Protocol #1 l Boom! studios Adventure Time Original Graphic Novel Volume 2: Pixel Princesses l Boom! studios 1 Ash & the Army of Darkness #1 l d. E./dynAmitE EntErtAinmEnt 1 Noir #1 l d. E./dynAmitE EntErtAinmEnt Legends of Red Sonja #1 l d. E./dynAmitE EntErtAinmEnt Maria M. -

Lettera Apostolica “Patris Corde” Del Santo Padre Francesco In
N. 0645 Martedì 08.12.2020 Lettera Apostolica “Patris Corde” del Santo Padre Francesco in occasione del 150.mo anniversario della dichiarazione di San Giuseppe quale Patrono della Chiesa Universale Testo in lingua italiana Traduzione in lingua latina Traduzione in lingua francese Traduzione in lingua inglese Traduzione in lingua tedesca Traduzione in lingua spagnola Traduzione in lingua portoghese Traduzione in lingua polacca Traduzione in lingua araba Testo in lingua italiana LETTERA APOSTOLICA PATRIS CORDE DEL SANTO PADRE FRANCESCO IN OCCASIONE DEL 150° ANNIVERSARIO DELLA DICHIARAZIONE DI SAN GIUSEPPE 2 QUALE PATRONO DELLA CHIESA UNIVERSALE CON CUORE DI PADRE: così Giuseppe ha amato Gesù, chiamato in tutti e quattro i Vangeli «il figlio di Giuseppe».[1] I due Evangelisti che hanno posto in rilievo la sua figura, Matteo e Luca, raccontano poco, ma a sufficienza per far capire che tipo di padre egli fosse e la missione affidatagli dalla Provvidenza. Sappiamo che egli era un umile falegname (cfr Mt 13,55), promesso sposo di Maria (cfr Mt 1,18; Lc 1,27); un «uomo giusto» (Mt 1,19), sempre pronto a eseguire la volontà di Dio manifestata nella sua Legge (cfr Lc 2,22.27.39) e mediante ben quattro sogni (cfr Mt 1,20; 2,13.19.22). Dopo un lungo e faticoso viaggio da Nazaret a Betlemme, vide nascere il Messia in una stalla, perché altrove «non c’era posto per loro» (Lc 2,7). Fu testimone dell’adorazione dei pastori (cfr Lc 2,8-20) e dei Magi (cfr Mt 2,1-12), che rappresentavano rispettivamente il popolo d’Israele e i popoli pagani. -

NEW THIS WEEK from DC... Legion of Super Heroes #3 Flash Forward
NEW THIS WEEK FROM DC... Legion of Super Heroes #3 Flash Forward #5 (of 6) Batman's Grave #4 (of 12) Flash #86 Teen Titans #38 Nightwing #68 Question the Death of Vic Sage #2 (of 4) Crisis on Infinite Earths Giant #1 Low Low Woods #2 (of 6) Aquaman #56 Freedom Fighters #12 (of 12) He Man and the Masters of the Multiverse #3 (of 6) Superman's Pal Jimmy Olden #7 (of 12) Justice League Odyssey #17 Lucifer #16 RWBY #4 (of 7) Detective Comics #359 Facsimile Edition Dollar Comics Batman Adventures #12 (1st Harley Quinn) Green Lantern Legacy GN Birds of Prey Black Canary GN NEW THIS WEEK FROM MARVEL... Avengers #29 Iron Man 2020 #1 (of 6) Revenge of the Cosmic Ghost Rider #2 (of 5) Venom the End Jessica Jones Blind Spot #1 (of 6) Valkyrie Jane Foster #7 Marvel's Black Widow Prelude #1 (of 2) Marvel's Spider-Man Black Cat Strikes #1 (of 5) Ruins of Ravencroft: Sabretooth Runaways #29 Incredible Hulk #180 Facsimile Edition Marvel Tales Ravencroft #1 True Believers Criminally Insane Mandarin True Believers Criminally Insane Gypsy Moth Amazing Spider-Man "Maximum Carnage" Epic Collection GN NEW THIS WEEK FROM IMAGE... Undiscovered Country #3 Dead Eyes #4 SFSX Safe Sex #5 Spawn #304 Hit Girl Season Two #12 Lucy Claire Redemption #2 Pretty Violent #6 ALSO NEW THIS WEEK! Red Mother #2 Skulldigger & Skeleton Boy #2 (of 6) Second Coming #6 (of 6) Bloodshot #5 Five Years #7 Hellboy Winter Special 2019 Rai #3 Heavy Metal #296 Machine Girl #3 Over The Ropes #2 (of 5) Rising Sun #1 Strangelands #5 Tales from Harrow County Death's Choir #2 (of 4) Vampirella Red Sonja #5 Comic Quest Buried City Elfquest Stargazers Hunt #2 (of 6) Forever Maps GN Ghosted in LA #7 Illustrators Special #5 Midnight Vista #5 Steeple #5 (of 5) NEW POP CULTURE.. -

Newsletter Term 2 2021
Newsletter Term 2 2021 Term 3 Date Claimers Principal’s Report Greetings to you all and Term 3 Dates 12/07/21 – 17/09/21 welcome to our term two newsletter. This edition is larger P&C Meeting 19/07/21 23/08/21 than a puppy’s love as it bulges with the achievements of our Cairns Show students across all aspects of Public Holiday 16/07/21 our school. Also contained in this publication is an update on our improvement agenda, the Mini School Tablelands 03/08/21 – 05/08/21 envisaging process, our staff Cairns 17/08/21 – 19/08/21 news and the Cairns to Karumba Peninsula 17/08/21 – 19/08/21 Bike Ride. Hospitality in Practice 16/08/21 – 20/08/21 School Improvement We continue to refine our practices regarding our 2021 Year 11 &12 areas of improvement: • Student Free Day 03/09/21 Developing viable pathways for our students in further education, training or employment STEAM Camp 01/09/21 – 02/09/21 • Implementing a whole of school moderation 09/09/21 – 10/09/21 process • Providing timely, specific, understandable and Music Camp 13/09/21 – 14/09/21 actionable feedback to our students and home tutors. Please send any feedback that you have regarding this process to the Executive Services Team via [email protected]. I look forward to reading your feedback on our progress. Our drive at Cairns SDE is to ensure our students have a safe place to learn about our world, so that they can take their place in the world. -

Today Your Barista Is: Genre Characteristics in the Coffee Shop Alternate Universe
Today Your Barista Is: Genre Characteristics in The Coffee Shop Alternate Universe Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Katharine Elizabeth McCain Graduate Program in English The Ohio State University 2020 Dissertation Committee Sean O’Sullivan, Advisor Matthew H. Birkhold Jared Gardner Elizabeth Hewitt 1 Copyright by Katharine Elizabeth McCain 2020 2 Abstract This dissertation, Today Your Barista Is: Genre Characteristics in The Coffee Shop Alternate Universe, works to categorize and introduce a heretofore unrecognized genre within the medium of fanfiction: The Coffee Shop Alternate Universe (AU). Building on previous sociological and ethnographic work within Fan Studies, scholarship that identifies fans as transformative creators who use fanfiction as a means of promoting progressive viewpoints, this dissertation argues that the Coffee Shop AU continues these efforts within a defined set of characteristics, merging the goals of fanfiction as a medium with the specific goals of a genre. These characteristics include the Coffee Shop AU’s structure, setting, archetypes, allegories, and the remediation of related mainstream genres, particularly the romantic comedy. The purpose of defining the Coffee Shop AU as its own genre is to help situate fanfiction within mainstream literature conventions—in as much as that’s possible—and laying the foundation for future close reading. This work also helps to demonstrate which characteristics are a part of a communally developed genre as opposed to individual works, which may assist in legal proceedings moving forward. However, more crucially this dissertation serves to encourage the continued, formal study of fanfiction as a literary and cultural phenomenon, one that is beginning to closely analyze the stories fans produce alongside the fans themselves. -

Annual Report 2013 For
Leelanau Conservancy Conserving the Land, Water and Scenic Character of Leelanau County 2013 Annual Report and Spring 2014 Newsletter Leelanau Forever: Protecting This Place We Love n late November, the Leelanau Enterprise featured our IClay Cliffs Natural Area project on its front page. The Conservancy, along with Leland Township, had just finalized the purchase of this unbelievably beautiful 104-acre property, with 1,700 feet of frontage on both Lake Michigan and Lake Leelanau. We loved the Enterprise story headline which shouted, “Ours!” along with a beautiful photo. When you think about it, the word “Ours” pretty much sums up all of the natural areas we have preserved for public enjoyment. Houdek Dunes, DeYoung, Chippewa Run, the Lake Leelanau Narrows, Swanson Preserve, Crystal River just to name a few…and now, Clay Cliffs. These special places are all collectively “Ours.” They are ours to enjoy, to find wonder in, to explore with our families. In our 25-year history, we have also protected thousands of acres of private lands and working farms with a legal tool called a conservation easement. We can’t call these places “Ours,” but there’s no denying that everyone who loves Leelanau is the beneficiary when important farm and natural lands are protected. Private protected lands give us some of the most stunning roadside views, they protect our water quality and wildlife, and they produce the local foods we all adore. Finalizing the Clay Cliffs purchase was just a part of a record-breaking 25th Anniversary year. In 2013 we protected an impressive 912 acres, bringing our 25-year total to nearly 11,000 acres. -

Is Infantilization Ethical? an Ethical Question for Gerontologists
Editorial: Is Infantilization Ethical? An Ethical Question for Gerontologists Stephen M. Marson, Ph.D., Senior Editor Journal of Social Work Values and Ethics, Volume 10, Number 1 (2013) Copyright 2013, Association of Social Work Boards (ASWB) This text may be freely shared among individuals, but it may not be republished in any medium without express written consent from the authors and advance notification ofASWB Since 1982 I have been observing health-care abuse. The critical issues are threefold: 1) professionals, including social workers, interact professionals are currently interacting with elderly with elderly residents in a syrupy and infantile persons in an infantile manner; 2) professionals manner. As a supervisor I never felt comfortable do not believe this style is harmful, but rather a with this type of interaction—but at the time—I demonstration of warmth; and 3) infantilization is did not have the skills to wrap coherent words harmful whether it is intentional or unintentional. around the behavior. As a doctoral student and a I have been asking policymakers in my state, practicing social worker I made a Herculean effort “Does infantilization of elderly persons constitute to conceptualize the interaction within a theoretical elder abuse?” The reply: “This is a difficult framework. I thought that if I had a theory I question to answer.” would have the effective words to modify the interaction I was observing in nursing homes. The bottom line is this: Professionals who are interacting with elderly persons in a syrupy and My first effort was to employ Goffman’s infantile manner have good intentions. On a Dramaturgical and Framework Analysis. -
![EAN Ref. M [G] EAN 02A010 1000 0/12 5902062030627 02A665 500 6/24 5902062030153 02A012 1250 0/12 5902062030320](https://docslib.b-cdn.net/cover/9083/ean-ref-m-g-ean-02a010-1000-0-12-5902062030627-02a665-500-6-24-5902062030153-02a012-1250-0-12-5902062030320-3499083.webp)
EAN Ref. M [G] EAN 02A010 1000 0/12 5902062030627 02A665 500 6/24 5902062030153 02A012 1250 0/12 5902062030320
Młotek stolarski / Claw hammer / Tischlerhammer / Молоток столярный / Молоток столярський / Szeghúzókalapács / ciocan Młotek murarski / Masonry hammer / Maurerhammer / Молоток каменщика / Молоток мулярський / Kőműves kalapács / ciocan tîmplar / Truhlářské kladívko / Kladivo stolárske / Tesarsko kladivo / Plaktukas staliui / Āmurs galdnieka / Sõrghaamer / Чук кози zidărie / Zednické kladívko / Kladivo murárske / Zidarsko kladivo / Plaktukas mūrininkui / Āmurs mūrnieka / Müürivasar / Чук крак / Stolarski čekić / Cekic / Σφυρί ξυλουργού / Martillo de carpintero / Martello di falegname / Martelo p. madeira / Martelet / Zidarski čekić / Cekic / Σφυρί κτίστη / Piqueta de albañil / Martello di muratore / Martelo p. Alvenaria / Martelet de maçons / de menuisier / Tanghamer Metselaarshamer Ref. m [g] EAN 02A911 450 6/36 5902062012555 Młotek ciesielski / Carpenter’s hammer / Zimmererhammer / Молоток шиферный / Молоток теслярський / Ácskalapács / Ciocan dulgerie / Tesařské kladívo / Tesárske kladivko / Mizarsko kladivo / Plaktukas dailidei / Namdara āmurs / Tislerihaamer / Дърводелски чук / Tesarski čekić / Stolarski čekić / Σφυρί πέτρας / Martillo de carpintero / Martello da carpentiere / Martelo de marcenaria / Martelet de charpentier / Timmermanshamer Ref. m [g] EAN 02A913 600 6/24 5902062012579 Młotek ślusarski / Machinist hammer / Schlosserhammer / Молоток слесарный / Молоток слюсарний / Kalapács / Młotek ślusarski / Zámečnické kladívko / Kladivo zámočnícke / Ključavničarsko kladivo / Plaktukas šaltkalviui / Āmurs atslēdznieka / Lukksepahaamer