Constitutive GLI1 Expression in Chondrosarcoma Is Regulated by Major Vault Protein Via Mtor/S6K1 Signaling Cascade
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Differential RNA Packaging Into Small Extracellular Vesicles by Neurons
Luo et al. Cell Commun Signal (2021) 19:75 https://doi.org/10.1186/s12964-021-00757-4 RESEARCH Open Access Diferential RNA packaging into small extracellular vesicles by neurons and astrocytes Xuan Luo1, Renée Jean‑Toussaint1, Ahmet Sacan2 and Seena K. Ajit1* Abstract Background: Small extracellular vesicles (sEVs) mediate intercellular communication by transferring RNA, proteins, and lipids to recipient cells. These cargo molecules are selectively loaded into sEVs and mirror the physiological state of the donor cells. Given that sEVs can cross the blood–brain barrier and their composition can change in neurologi‑ cal disorders, the molecular signatures of sEVs in circulation can be potential disease biomarkers. Characterizing the molecular composition of sEVs from diferent cell types is an important frst step in determining which donor cells contribute to the circulating sEVs. Methods: Cell culture supernatants from primary mouse cortical neurons and astrocytes were used to purify sEVs by diferential ultracentrifugation and sEVs were characterized using nanoparticle tracking analysis, transmission electron microscopy and western blot. RNA sequencing was used to determine diferential expression and loading patterns of miRNAs in sEVs released by primary neurons and astrocytes. Motif analysis was conducted on enriched miRNAs in sEVs and their respective donor cells. Results: Sequencing total cellular RNA, and miRNAs from sEVs isolated from culture media of postnatal mouse corti‑ cal neurons and astrocytes revealed a distinct profle between sEVs and their corresponding cells. Though the total number of detected miRNAs in astrocytes was greater than neurons, neurons expressed more sEV‑associated miRNAs than astrocytes. Only 20.7% of astrocytic miRNAs were loaded into sEVs, while 41.0% of neuronal miRNAs were loaded into sEVs, suggesting diferences in the cellular sorting mechanisms. -

Novel Mechanisms of Pten Dysfunction in Pten Hamartoma Tumor Syndromes
NOVEL MECHANISMS OF PTEN DYSFUNCTION IN PTEN HAMARTOMA TUMOR SYNDROMES DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Marcus G. Pezzolesi, B.S. ***** The Ohio State University 2008 Dissertation Committee: Approved by Professor Allan J. Yates, Advisor Professor Charis Eng, Co-Advisor _________________________________ Professor Wolfgang Sadee Advisor Integrated Biomedical Science Professor Michael C. Ostrowski Graduate Program Professor Lawrence S. Kirschner Professor Lei Shen ABSTRACT Phosphatase and tensin homolog deleted on chromosome ten (PTEN) encodes a tumor suppressor phosphatase frequently mutated in both sporadic and heritable forms of human cancer. Germline mutations in PTEN are associated with a number of heritable cancer syndromes referred to as the PTEN hamartoma tumor syndromes (PHTS) and includes both Cowden syndrome (CS) and Bannayan-Riley-Ruvalcaba syndrome (BRRS). Data from our laboratory suggests that alternate mechanisms of PTEN deregulation are likely to, at least in part, contribute to dysfunction in patients with these syndromes, particularly in those for whom germline mutations have yet to be identified. To better understand the mechanism(s) underlying dysregulation of PTEN in these syndromes, we employed a series of genetic and biochemical approaches aimed at investigating novel mechanisms involved in the regulation and deregulation of PTEN. Using a haplotype-based approach, we identified specific haplotypes and rare alleles within the PTEN locus that contribute to disease susceptibility and the phenotypic complexity of this syndrome. Within a haplotype block associated with PTEN-mutation negative patients, we identified a canonical E-box sequence located upstream of PTEN’s minimal promoter. -

Major Vault Protein, a Candidate Gene in 16P11.2 Microdeletion Syndrome, Is Required for the Homeostatic Regulation of Visual Cortical Plasticity
This Accepted Manuscript has not been copyedited and formatted. The final version may differ from this version. Research Articles: Development/Plasticity/Repair Major vault protein, a candidate gene in 16p11.2 microdeletion syndrome, is required for the homeostatic regulation of visual cortical plasticity Jacque P K Ip1, Ikue Nagakura1, Jeremy Petravicz1, Keji Li1, Erik A.C. Wiemer2 and Mriganka Sur1 1Department of Brain and Cognitive Sciences, Picower Institute for Learning and Memory, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, Massachusetts 02139 USA 2Department of Medical Oncology, Erasmus MC Cancer Institute, Erasmus University Medical Center, Wytemaweg 80, 3015 CN Rotterdam, The Netherlands DOI: 10.1523/JNEUROSCI.2034-17.2018 Received: 18 July 2017 Revised: 17 February 2018 Accepted: 2 March 2018 Published: 14 March 2018 Author contributions: K.L. edited the paper; J.P.K.I., I.N., and M.S. designed research; J.P.K.I., I.N., J.P., and K.L. performed research; E.A.W. contributed unpublished reagents/analytic tools; J.P.K.I., I.N., J.P., and K.L. analyzed data; J.P.K.I., I.N., and M.S. wrote the paper. Conflict of Interest: The authors declare no competing financial interests. This work was supported by Human Frontier Science Program Long-Term Fellowship (J.P.K.I), NIH grants MH085802 and EY007023 (M.S.), Simons Postdoctoral Fellowship (I.N.) and the Simons Foundation Autism Research Initiative through the Simons Center for the Social Brain, MIT (M.S.). We thank Bess Rosen for technical assistance. Correspondence: Mriganka Sur ([email protected]) Department of Brain and Cognitive Sciences, Picower Institute for Learning and Memory, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, Massachusetts 02139 USA Cite as: J. -

The Malignant Role of Exosomes As Nanocarriers of Rare RNA Species
International Journal of Molecular Sciences Review The Malignant Role of Exosomes as Nanocarriers of Rare RNA Species Alina-Andreea Zimta 1 , Olafur Eysteinn Sigurjonsson 2,3 , Diana Gulei 1,* and Ciprian Tomuleasa 1,4 1 Research Center for Advanced Medicine-Medfuture, Iuliu Hatieganu University of Medicine and Pharmacy, 400012 Cluj-Napoca, Romania; [email protected] (A.-A.Z.); [email protected] (C.T.) 2 The Blood Bank, Landspitali University Hospital, 121 Reykjavik, Iceland; [email protected] 3 School of Science and Engineering, Reykjavik University, 107 Reykjavik, Iceland 4 Department of Hematology, Oncology Institute Prof. Dr. Ion Chiricuta, 400015 Cluj-Napoca, Romania * Correspondence: [email protected] or [email protected] Received: 10 July 2020; Accepted: 13 August 2020; Published: 15 August 2020 Abstract: Nowadays, advancements in the oncology sector regarding diagnosis methods allow us to specifically detect an increased number of cancer patients, some of them in incipient stages. However, one of the main issues consists of the invasive character of most of the diagnosis protocols or complex medical procedures associated with it, that impedes part of the patients to undergo routine checkups. Therefore, in order to increase the number of cancer cases diagnosed in incipient stages, other minimally invasive alternatives must be considered. The current review paper presents the value of rare RNA species isolated from circulatory exosomes as biomarkers of diagnosis, prognosis or even therapeutic intervention. Rare RNAs are most of the time overlooked in current research in favor of the more abundant RNA species like microRNAs. However, their high degree of stability, low variability and, for most of them, conservation across species could shift the interest toward these types of RNAs. -

A Master Autoantigen-Ome Links Alternative Splicing, Female Predilection, and COVID-19 to Autoimmune Diseases
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.30.454526; this version posted August 4, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. A Master Autoantigen-ome Links Alternative Splicing, Female Predilection, and COVID-19 to Autoimmune Diseases Julia Y. Wang1*, Michael W. Roehrl1, Victor B. Roehrl1, and Michael H. Roehrl2* 1 Curandis, New York, USA 2 Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, USA * Correspondence: [email protected] or [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.30.454526; this version posted August 4, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Chronic and debilitating autoimmune sequelae pose a grave concern for the post-COVID-19 pandemic era. Based on our discovery that the glycosaminoglycan dermatan sulfate (DS) displays peculiar affinity to apoptotic cells and autoantigens (autoAgs) and that DS-autoAg complexes cooperatively stimulate autoreactive B1 cell responses, we compiled a database of 751 candidate autoAgs from six human cell types. At least 657 of these have been found to be affected by SARS-CoV-2 infection based on currently available multi-omic COVID data, and at least 400 are confirmed targets of autoantibodies in a wide array of autoimmune diseases and cancer. -

A Chronic Hypoxic Response in Photoreceptors Alters the Vitreous Proteome in Mice
Research Collection Journal Article A chronic hypoxic response in photoreceptors alters the vitreous proteome in mice Author(s): Schori, Christian; Trachsel, Christian; Grossmann, Jonas; Barben, Maya; Klee, Katrin; Storti, Federica; Samardzija, Marijana; Grimm, Christian Publication Date: 2019-08 Permanent Link: https://doi.org/10.3929/ethz-b-000348912 Originally published in: Experimental eye research 185, http://doi.org/10.1016/j.exer.2019.107690 Rights / License: Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International This page was generated automatically upon download from the ETH Zurich Research Collection. For more information please consult the Terms of use. ETH Library Experimental Eye Research 185 (2019) 107690 Contents lists available at ScienceDirect Experimental Eye Research journal homepage: www.elsevier.com/locate/yexer A chronic hypoxic response in photoreceptors alters the vitreous proteome in mice T Christian Schoria,b, Christian Trachselc, Jonas Grossmannc, Maya Barbena,d, Katrin Kleea,b, ∗ Federica Stortia, Marijana Samardzijaa, Christian Grimma,b,d, a Lab for Retinal Cell Biology, Dept. Ophthalmology, University of Zurich, Zurich, Switzerland b Center for Integrative Human Physiology (ZIHP), University of Zurich, Zurich, Switzerland c Functional Genomics Center Zurich (FGCZ), ETH Zurich and University of Zurich, Zurich, Switzerland d Neuroscience Center Zurich (ZNZ), University of Zurich, Zurich, Switzerland ARTICLE INFO ABSTRACT Keywords: Reduced oxygenation of the outer retina in the aging eye may activate a chronic hypoxic response in RPE and Vitreous photoreceptor cells and is considered as a risk factor for the development of age-related macular degeneration Hypoxia (AMD). In mice, a chronically active hypoxic response in the retinal pigment epithelium (RPE) or photoreceptors Proteomics leads to age-dependent retinal degeneration. -

Expression Patterns of the Major Vault Protein (MVP) and Cellular Vault Particles in Aquatic Animal Models Alyssa L
Clemson University TigerPrints All Theses Theses 5-2016 Expression Patterns of the Major Vault Protein (MVP) and Cellular Vault Particles in Aquatic Animal Models Alyssa L. Margiotta Clemson University, [email protected] Follow this and additional works at: https://tigerprints.clemson.edu/all_theses Recommended Citation Margiotta, Alyssa L., "Expression Patterns of the Major Vault Protein (MVP) and Cellular Vault Particles in Aquatic Animal Models" (2016). All Theses. 2391. https://tigerprints.clemson.edu/all_theses/2391 This Thesis is brought to you for free and open access by the Theses at TigerPrints. It has been accepted for inclusion in All Theses by an authorized administrator of TigerPrints. For more information, please contact [email protected]. EXPRESSION PATTERNS OF THE MAJOR VAULT PROTEIN (MVP) AND CELLULAR VAULT PARTICLES IN AQUATIC ANIMAL MODELS A Thesis Presented to the Graduate School of Clemson University In Partial Fulfillment of the Requirements for the Degree Master of Science Biological Sciences by Alyssa L. Margiotta May 2016 Accepted by: Charles D. Rice, Ph.D., Committee Chair Yanzhang Wei, Ph.D. Thomas R. Scott, Ph.D. ABSTRACT Cellular vaults are ubiquitous 13 mega Da multi-subunit structures that may have a role in nucleo-cytoplasmic transport. Seventy percent of the vault's mass consists of a ≈100 kDa protein, the major vault protein (MVP). Elevated MVP was first recognized as lung resistance protein (LRP) because metastatic lymphoid tumor cells in the lung over- expressed this protein following acquired resistance to traditional chemotherapy. Previous work in our lab screened a cDNA library constructed from channel catfish monocytes (42TA cells), whereby MVP was sequenced and found to be highly conserved compared to other vertebrates. -

Proteomic Study to Survey the CIGB-552 Antitumor Effect
Hindawi Publishing Corporation BioMed Research International Volume 2015, Article ID 124082, 18 pages http://dx.doi.org/10.1155/2015/124082 Research Article Proteomic Study to Survey the CIGB-552 Antitumor Effect Arielis Rodríguez-Ulloa,1 Jeovanis Gil,1 Yassel Ramos,1 Lilian Hernández-Álvarez,2 Lisandra Flores,1 Brizaida Oliva,3 Dayana García,1 Aniel Sánchez-Puente,1 Alexis Musacchio-Lasa,2 Jorge Fernández-de-Cossio,2 Gabriel Padrón,1 Luis J. González López,1 Vladimir Besada,1 and Maribel Guerra-Vallespí3 1 DepartmentofProteomics,CenterforGeneticEngineeringandBiotechnology,10600Havana,Cuba 2Department of Bioinformatics, Center for Genetic Engineering and Biotechnology, 10600 Havana, Cuba 3Pharmaceutical Department, Center for Genetic Engineering and Biotechnology, 10600 Havana, Cuba Correspondence should be addressed to Arielis Rodr´ıguez-Ulloa; [email protected] Received 11 March 2015; Accepted 26 August 2015 Academic Editor: Zheng Li Copyright © 2015 Arielis Rodr´ıguez-Ulloa et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. CIGB-552 is a cell-penetrating peptide that exerts in vitro and in vivo antitumor effect on cancer cells. In the present work, the mechanism involved in such anticancer activity was studied using chemical proteomics and expression-based proteomics in culture cancer cell lines. CIGB-552 interacts with at least 55 proteins, as determined by chemical proteomics. A temporal differential proteomics based on iTRAQ quantification method was performed to identify CIGB-552 modulated proteins. The proteomic profile includes 72 differentially expressed proteins in response to CIGB-552 treatment. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -
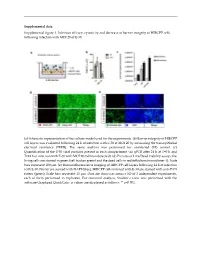
Supplemental Figure 1. Infection Efficacy, Cytoxicity and Decrease in Barrier Integrity of HIBCPP Cells Following Infection with MOI 20 of E-30
Supplemental data: Supplemental figure 1. Infection efficacy, cytoxicity and decrease in barrier integrity of HIBCPP cells following infection with MOI 20 of E-30. (a) Schematic representation of the culture model used for the experiments. (b) Barrier integrity of HIBCPP cell layers was evaluated following 24 h of infection with E-30 at MOI 20 by measuring the transepithelial electrical resistance (TEER). The same analysis was performed for uninfected (UI) control. (c) Quantification of the E-30 viral particles present in each compartment via qPCR after 24 h at T=0 h and T=24 h of infection with E-30 with MOI 20 (nd=non detected). (d) Pictures of Live/Dead viability assays; the living cells are stained in green (cell tracker green) and the dead cells in red (ethidium homodimer-1). Scale bars represent 100 µm. (e) Immunofluorescence imaging of HIBCPP cell layers following 24 h of infection with E-30. Nuclei are stained with DAPI (blue), HIBCPP cells infected with E-30 are stained with anti-PAN entero (green). Scale bars represent 15 µm. Data are shown as mean ± SD of 3 independent experiments, each of them performed in triplicates. For statistical analysis, Student’s t-test was performed with the software Graphpad QuickCalcs. p values are displayed as follows: ** p<0.001. Supplemental table 1. Up-regulated proteins in the lipid raft of HIBCPP cells following 24 h of E-30 infection at MOI 20. Proteins were identified as significantly changed in abundancy, when they exceeded a t-test difference > 1.0. Results shown are the comparison of 3 different independent proteomic experiments. -

The Endosome Is a Master Regulator of Plasma Membrane Collagen Fibril Assembly
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.25.436925; this version posted March 25, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The endosome is a master regulator of plasma membrane collagen fibril assembly 1Joan Chang*, 1Adam Pickard, 1Richa Garva, 1Yinhui Lu, 2Donald Gullberg and 1Karl E. Kadler* 1Wellcome Centre for Cell-Matrix Research, Faculty of Biology, Medical and Health, University of Manchester, Michael Smith Building, Oxford Road, Manchester M13 9PT UK, 2Department of Biomedicine and Center for Cancer Biomarkers, Norwegian Center of Excellence, University of Bergen, Norway. * Co-corresponding authors: JC email: [email protected] (orcid.org/0000-0002-7283- 9759); KEK email: [email protected] (orcid.org/0000-0003-4977-4683) Keywords: collagen-I, endocytosis, extracellular matrix, fibril, fibrillogenesis, integrin-a11, trafficking, VPS33b, [abstract] [149 word max] Collagen fibrils are the principal supporting elements in vertebrate tissues. They account for 25% of total protein mass, exhibit a broad range of size and organisation depending on tissue and stage of development, and can be under circadian clock control. Here we show that the remarkable dynamic pleomorphism of collagen fibrils is underpinned by a mechanism that distinguishes between collagen secretion and initiation of fibril assembly, at the plasma membrane. Collagen fibrillogenesis occurring at the plasma membrane requires vacuolar protein sorting (VPS) 33b (which is under circadian clock control), collagen-binding integrin-a11 subunit, and is reduced when endocytosis is inhibited. Fibroblasts lacking VPS33b secrete soluble collagen without assembling fibrils, whereas constitutive over-expression of VPS33b increases fibril number with loss of fibril rhythmicity. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses by a Synthetic TLR3 Ligand Mitigates
Downloaded from http://www.jimmunol.org/ by guest on September 28, 2021 Inducing is online at: average * The Journal of Immunology published online 16 August 2013 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2013/08/16/jimmun ol.1300376 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 28, 2021. Published August 16, 2013, doi:10.4049/jimmunol.1300376 The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN.