Unveiling Role of Sphingosine-1-Phosphate Receptor 2 As a Brake of Epithelial Stem Cell Proliferation and a Tumor Suppressor In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mutant Luteinizing Hormone Receptors in a Compound Heterozygous Patient with Complete Leydig Cell Hypoplasia: Abnormal Processing Causes Signaling Deficiency
0013-7227/02/$15.00/0 The Journal of Clinical Endocrinology & Metabolism 87(6):2506–2513 Printed in U.S.A. Copyright © 2002 by The Endocrine Society Mutant Luteinizing Hormone Receptors in a Compound Heterozygous Patient with Complete Leydig Cell Hypoplasia: Abnormal Processing Causes Signaling Deficiency J. W. M. MARTENS, S. LUMBROSO, M. VERHOEF-POST, V. GEORGET, A. RICHTER-UNRUH, M. SZARRAS-CZAPNIK, T. E. ROMER, H. G. BRUNNER, A. P. N. THEMMEN, AND CH. SULTAN Departments of Endocrinology and Reproduction and Internal Medicine, Erasmus University (J.W.M.M., M.V.-P., A.P.N.T.), 3000 DR Rotterdam, The Netherlands; Hormonologie du De´veloppement et de la Reproduction, Hoˆpital Lapeyronie and INSERM, U-439 (S.L., V.G., C.S.), 34090 Montpellier, France; Endocrinologie et Gyne´cologie Pe´diatriques, Hoˆpital A. de Villeneuve (C.S.), 34295 Montpellier, France; Department of Pediatric Endocrinology, University Children’s Hospital, University of Essen (A.R.-U.), 45122 Essen, Germany; Department of Pediatric Endocrinology, Children’s Memorial Health Institute (M.S.-C., T.E.R.), 04-730 Warsaw, Poland; and Department of Human Genetics, University Hospital (H.G.B.), 6500 HB Nijmegen, The Netherlands Over the past 5 yr several inactivating mutations in the absence of total and cell surface hormone binding, protein LH receptor gene have been demonstrated to cause Leydig levels of both mutant LH receptors are only moderately af- cell hypoplasia, a rare autosomal recessive form of male fected. The expression and study of enhanced green fluores- pseudohermaphroditism. Here, we report the identification of cent protein-tagged receptors confirmed this view and fur- two new LH receptor mutations in a compound heterozygous ther indicated that initial translocation to the endoplasmic case of complete Leydig hypoplasia and determine the cause reticulum of these mutant receptors is normal. -

LGR5 and LGR6 in Stem Cell Biology and Ovarian Cancer
www.impactjournals.com/oncotarget/ Oncotarget, 2018, Vol. 9, (No. 1), pp: 1346-1355 Review LGR5 and LGR6 in stem cell biology and ovarian cancer Adam J. Schindler1, Arisa Watanabe1 and Stephen B. Howell1 1Moores Cancer Center, University of California, San Diego, CA, USA Correspondence to: Stephen B. Howell, email: [email protected] Keywords: ovarian cancer, Wnt, LGR6, LGR5, RSPO Received: June 14, 2017 Accepted: July 31, 2017 Published: August 11, 2017 Copyright: Schindler et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Wnt signaling plays a fundamental role in patterning of the embryo and maintenance of stem cells in numerous epithelia. Epithelial stem cells are closeted in niches created by surrounding differentiated cells that express secreted Wnt and R-spondin proteins that influence proliferation rate and fate determination of stem cell daughters. R-spondins act through the LGR receptors to enhance Wnt signaling. This close association of stem cells with more differentiated regulatory cells expressing Wnt-pathway ligands is a feature replicated in all of the epithelial stem cell systems thus far examined. How the stem cell niche operates through these short-range interactions is best understood for the crypts of the gastrointestinal epithelium and skin. Less well understood are the stem cells that function in the ovarian surface epithelium (OSE) and fallopian tube epithelium (FTE). While the cuboidal OSE appears to be made up of a single cell type, the cells of the FTE progress through a life cycle that involves differentiation into ciliated and secretory subtypes that are eventually shed into the lumen in a manner similar to the gastrointestinal epithelium. -

Kynurenine Promotes the Goblet Cell Differentiation of HT-29 Colon Carcinoma Cells by Modulating Wnt, Notch and Ahr Signals
1930 ONCOLOGY REPORTS 39: 1930-1938, 2018 Kynurenine promotes the goblet cell differentiation of HT-29 colon carcinoma cells by modulating Wnt, Notch and AhR signals JOO-HUNG PARK, JEONG-MIN LEE, EUN-JIN LEE, DA-JEONG KIM and WON-BHIN Hwang Department of Biology, Changwon National University, Changwon, Kyungnam 51140, Republic of Korea Received August 31, 2017; Accepted February 7, 2018 DOI: 10.3892/or.2018.6266 Abstract. Various amino acids regulate cell growth and inorganic salts for normal cell metabolism. However, the differentiation. In the present study, we examined the ability composition of medium formulations vary widely in concen- of HT-29 cells to differentiate into goblet cells in RPMI trations of glucose and other metabolic precursors, including and DMEM which are largely different in the amounts of amino acids. Thus, many types of cells showed different numerous amino acids. Most of the HT-29 cells differentiated responses to culture media in regards to proliferation and into goblet cells downregulating the stem cell marker Lgr5 differentiation. Th17 differentiation was induced more effi- when cultured in DMEM, but remained undifferentiated in ciently in Iscove's modified Dulbecco's medium (IMDM) than RPMI. The goblet cell differentiation in DMEM was inhibited in RPMI (1). Bone marrow-derived dendritic cells in IMDM by 1-methyl-tryptophan (1-MT), an inhibitor of indoleamine expressed higher levels of co-stimulatory and MHC II mole- 2,3 dioxygenase-1 which is the initial enzyme in tryptophan cules, compared to the cells generated in RPMI (2). In addition, metabolism along the kynurenine (KN) pathway, whereas hepatocyte differentiation and propagation and phenotype of tryptophan and KN induced goblet cell differentiation in corneal stroma-derived stem cells were modulated by culture RPMI. -

CV Francesco Patti Name Francesco Patti Born in Acireale (Italy) 12/06/1958 Scholastic Career II Level Education 1976 Degree Me
Name Francesco Patti Born in Acireale (Italy) 12/06/1958 Scholastic career II level education 1976 Degree Medicine and Surgery 1982 maximum cum laude Post Degree Neurology, University of Catania, 1986 Post Degree Phyisiotherapy, University of Parma, 1990 Work activity 1987-1988 Regional fellowship as Junior Neurologist to study “ Descriptive Neuroepidemiology of most frequent neurological diseases in Sicily”, progetto Regionale 55/P, sponsored by WHO. 1987–1989 Assisting Professor of Neuroendocrinology and Neuroimmunology Scuola di Specializzazione in Neurologia, University of Catania 1991–2000 “CollaboratoreTecnico” Chair of Neurorheabilitation, Institute of Neurological Sciences, University of Catania. 2000–2002 “Tecnico Laureato” (Funzionario Tecnico) Department of Neurological Sciences November 2002 – October 2014 “Ricercatore Confermato” (Aggregate Professor) Clinical Researcher Department of Neurological Sciences, University of Catania. November 2014 - up till now Associate Professor of Neurology, Department of Medical and Surgical Sciences and Advanced Tecnologies G.F. Ingrassia, University of Catania. Clinician profile and activities He is responsible of the tertiary centre of multiple sclerosis at the University of Catania (Italy). The centre follows more than 2500 patients suffering from Multiple Sclerosis and few patients suffering from Devic Disease and Devic Spectrum disorder diseases. The centre follows also patients suffering from ALS (currently 100 patients) and other people with different forms of spasticity, offering them with a multidisciplinary approach every kind of Pag. 1 of 3 CV Francesco Patti You created this PDF from an application that is not licensed to print to novaPDF printer (http://www.novapdf.com) assistance. Scientific activity Research Interests ; Preclinical (1981-1988) ; Neurochemistry, Neuroendocrinolgy, Neuropsycopharmacology ; Clinical: (1989-current) ; Neuroepidemiology ; Clinical Immunology ; Quality of Life ; Neurorehabilitation ; Multiple Sclerosis. -

FTY720P Upregulates the Na+/K+ Atpase in Hepg2 Cells by Activating S1PR3 and Inducing PGE2 Release
Cellular Physiology Cell Physiol Biochem 2019;53:518-531 DOI: 10.33594/00000015510.33594/000000155 © 2019 The Author(s).© 2019 Published The Author(s) by and Biochemistry Published online: 11 SeptemberSeptember 20192019 Cell Physiol BiochemPublished Press GmbH&Co. by Cell Physiol KG Biochem 518 + + Press GmbH&Co. KG, Duesseldorf ChakkourAccepted: 5et September al.: FTY720-P 2019 Upregulates Na /K Atpase www.cellphysiolbiochem.com This article is licensed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 Interna- tional License (CC BY-NC-ND). Usage and distribution for commercial purposes as well as any distribution of modified material requires written permission. Original Paper FTY720P Upregulates the Na+/K+ ATPase in HepG2 Cells by Activating S1PR3 and Inducing PGE2 Release Mohamed Chakkour Sawsan Kreydiyyeh Department of Biology, Faculty of Arts & Sciences, American University of Beirut, Beirut, Lebanon Key Words FTY720-P • Na+/K+ATPase • HepG2 • PKC • ERK • PGE2 Abstract Background/Aims: Liver regeneration is induced by S1P and accompanied with an increase in hepatic Na+/K+ ATPase activity, suggesting a potential modulatory role of the sphingolipid on the ATPase activity. The ability of S1P to alter the ATPase activity was confirmed in a previous work which showed a time dependent effect, with an inhibition appearing at 15min and a stimulation at two hours. The aim of this work was to investigate if FTY720-P, an analogue of S1P used in the treatment of multiple sclerosis, exerts a similar effect at 2 hours. Methods: HepG2 cells were treated with FTY720-P for two hours and the activity of the Na+/K+ ATPase was assayed by measuring the amount of inorganic phosphate liberated in presence and absence of ouabain. -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -
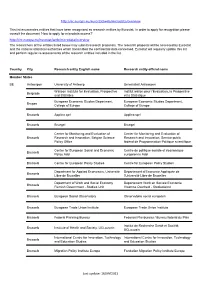
Eurostat: Recognized Research Entity
http://ec.europa.eu/eurostat/web/microdata/overview This list enumerates entities that have been recognised as research entities by Eurostat. In order to apply for recognition please consult the document 'How to apply for microdata access?' http://ec.europa.eu/eurostat/web/microdata/overview The researchers of the entities listed below may submit research proposals. The research proposal will be assessed by Eurostat and the national statistical authorities which transmitted the confidential data concerned. Eurostat will regularly update this list and perform regular re-assessments of the research entities included in the list. Country City Research entity English name Research entity official name Member States BE Antwerpen University of Antwerp Universiteit Antwerpen Walloon Institute for Evaluation, Prospective Institut wallon pour l'Evaluation, la Prospective Belgrade and Statistics et la Statistique European Economic Studies Department, European Economic Studies Department, Bruges College of Europe College of Europe Brussels Applica sprl Applica sprl Brussels Bruegel Bruegel Center for Monitoring and Evaluation of Center for Monitoring and Evaluation of Brussels Research and Innovation, Belgian Science Research and Innovation, Service public Policy Office fédéral de Programmation Politique scientifique Centre for European Social and Economic Centre de politique sociale et économique Brussels Policy Asbl européenne Asbl Brussels Centre for European Policy Studies Centre for European Policy Studies Department for Applied Economics, -

Lgr5 Homologues Associate with Wnt Receptors and Mediate R-Spondin Signalling
ARTICLE doi:10.1038/nature10337 Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling Wim de Lau1*, Nick Barker1{*, Teck Y. Low2, Bon-Kyoung Koo1, Vivian S. W. Li1, Hans Teunissen1, Pekka Kujala3, Andrea Haegebarth1{, Peter J. Peters3, Marc van de Wetering1, Daniel E. Stange1, Johan E. van Es1, Daniele Guardavaccaro1, Richard B. M. Schasfoort4, Yasuaki Mohri5, Katsuhiko Nishimori5, Shabaz Mohammed2, Albert J. R. Heck2 & Hans Clevers1 The adult stem cell marker Lgr5 and its relative Lgr4 are often co-expressed in Wnt-driven proliferative compartments. We find that conditional deletion of both genes in the mouse gut impairs Wnt target gene expression and results in the rapid demise of intestinal crypts, thus phenocopying Wnt pathway inhibition. Mass spectrometry demonstrates that Lgr4 and Lgr5 associate with the Frizzled/Lrp Wnt receptor complex. Each of the four R-spondins, secreted Wnt pathway agonists, can bind to Lgr4, -5 and -6. In HEK293 cells, RSPO1 enhances canonical WNT signals initiated by WNT3A. Removal of LGR4 does not affect WNT3A signalling, but abrogates the RSPO1-mediated signal enhancement, a phenomenon rescued by re-expression of LGR4, -5 or -6. Genetic deletion of Lgr4/5 in mouse intestinal crypt cultures phenocopies withdrawal of Rspo1 and can be rescued by Wnt pathway activation. Lgr5 homologues are facultative Wnt receptor components that mediate Wnt signal enhancement by soluble R-spondin proteins. These results will guide future studies towards the application of R-spondins for regenerative purposes of tissues expressing Lgr5 homologues. The genes Lgr4, Lgr5 and Lgr6 encode orphan 7-transmembrane 4–5 post-induction onwards. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Targeting Lysophosphatidic Acid in Cancer: the Issues in Moving from Bench to Bedside
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by IUPUIScholarWorks cancers Review Targeting Lysophosphatidic Acid in Cancer: The Issues in Moving from Bench to Bedside Yan Xu Department of Obstetrics and Gynecology, Indiana University School of Medicine, 950 W. Walnut Street R2-E380, Indianapolis, IN 46202, USA; [email protected]; Tel.: +1-317-274-3972 Received: 28 August 2019; Accepted: 8 October 2019; Published: 10 October 2019 Abstract: Since the clear demonstration of lysophosphatidic acid (LPA)’s pathological roles in cancer in the mid-1990s, more than 1000 papers relating LPA to various types of cancer were published. Through these studies, LPA was established as a target for cancer. Although LPA-related inhibitors entered clinical trials for fibrosis, the concept of targeting LPA is yet to be moved to clinical cancer treatment. The major challenges that we are facing in moving LPA application from bench to bedside include the intrinsic and complicated metabolic, functional, and signaling properties of LPA, as well as technical issues, which are discussed in this review. Potential strategies and perspectives to improve the translational progress are suggested. Despite these challenges, we are optimistic that LPA blockage, particularly in combination with other agents, is on the horizon to be incorporated into clinical applications. Keywords: Autotaxin (ATX); ovarian cancer (OC); cancer stem cell (CSC); electrospray ionization tandem mass spectrometry (ESI-MS/MS); G-protein coupled receptor (GPCR); lipid phosphate phosphatase enzymes (LPPs); lysophosphatidic acid (LPA); phospholipase A2 enzymes (PLA2s); nuclear receptor peroxisome proliferator-activated receptor (PPAR); sphingosine-1 phosphate (S1P) 1. -

A Benchmarking Study on Virtual Ligand Screening Against Homology Models of Human Gpcrs
bioRxiv preprint doi: https://doi.org/10.1101/284075; this version posted March 19, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A benchmarking study on virtual ligand screening against homology models of human GPCRs Victor Jun Yu Lima,b, Weina Dua, Yu Zong Chenc, Hao Fana,d aBioinformatics Institute (BII), Agency for Science, Technology and Research (A*STAR), 30 Biopolis Street, Matrix No. 07-01, 138671, Singapore; bSaw Swee Hock School of Public Health, National University of Singapore, 12 Science Drive 2, 117549, Singapore; cDepartment of Pharmacy, National University of Singapore, 18 Science Drive 4, 117543, Singapore; dDepartment of Biological Sciences, National University of Singapore, 16 Science Drive 4, Singapore 117558 To whom correspondence should be addressed: Dr. Hao Fan, Bioinformatics Institute (BII), Agency for Science, Technology and Research (A*STAR), 30 Biopolis Street, Matrix No. 07-01, 138671, Singapore; Telephone: +65 64788500; Email: [email protected] Keywords: G-protein-coupled receptor, GPCR, X-ray crystallography, virtual screening, homology modelling, consensus enrichment 1 bioRxiv preprint doi: https://doi.org/10.1101/284075; this version posted March 19, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract G-protein-coupled receptor (GPCR) is an important target class of proteins for drug discovery, with over 27% of FDA-approved drugs targeting GPCRs. However, being a membrane protein, it is difficult to obtain the 3D crystal structures of GPCRs for virtual screening of ligands by molecular docking. -

Supplementary Table S5. Differentially Expressed Gene Lists of PD-1High CD39+ CD8 Tils According to 4-1BB Expression Compared to PD-1+ CD39- CD8 Tils
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Immunother Cancer Supplementary Table S5. Differentially expressed gene lists of PD-1high CD39+ CD8 TILs according to 4-1BB expression compared to PD-1+ CD39- CD8 TILs Up- or down- regulated genes in Up- or down- regulated genes Up- or down- regulated genes only PD-1high CD39+ CD8 TILs only in 4-1BBneg PD-1high CD39+ in 4-1BBpos PD-1high CD39+ CD8 compared to PD-1+ CD39- CD8 CD8 TILs compared to PD-1+ TILs compared to PD-1+ CD39- TILs CD39- CD8 TILs CD8 TILs IL7R KLRG1 TNFSF4 ENTPD1 DHRS3 LEF1 ITGA5 MKI67 PZP KLF3 RYR2 SIK1B ANK3 LYST PPP1R3B ETV1 ADAM28 H2AC13 CCR7 GFOD1 RASGRP2 ITGAX MAST4 RAD51AP1 MYO1E CLCF1 NEBL S1PR5 VCL MPP7 MS4A6A PHLDB1 GFPT2 TNF RPL3 SPRY4 VCAM1 B4GALT5 TIPARP TNS3 PDCD1 POLQ AKAP5 IL6ST LY9 PLXND1 PLEKHA1 NEU1 DGKH SPRY2 PLEKHG3 IKZF4 MTX3 PARK7 ATP8B4 SYT11 PTGER4 SORL1 RAB11FIP5 BRCA1 MAP4K3 NCR1 CCR4 S1PR1 PDE8A IFIT2 EPHA4 ARHGEF12 PAICS PELI2 LAT2 GPRASP1 TTN RPLP0 IL4I1 AUTS2 RPS3 CDCA3 NHS LONRF2 CDC42EP3 SLCO3A1 RRM2 ADAMTSL4 INPP5F ARHGAP31 ESCO2 ADRB2 CSF1 WDHD1 GOLIM4 CDK5RAP1 CD69 GLUL HJURP SHC4 GNLY TTC9 HELLS DPP4 IL23A PITPNC1 TOX ARHGEF9 EXO1 SLC4A4 CKAP4 CARMIL3 NHSL2 DZIP3 GINS1 FUT8 UBASH3B CDCA5 PDE7B SOGA1 CDC45 NR3C2 TRIB1 KIF14 TRAF5 LIMS1 PPP1R2C TNFRSF9 KLRC2 POLA1 CD80 ATP10D CDCA8 SETD7 IER2 PATL2 CCDC141 CD84 HSPA6 CYB561 MPHOSPH9 CLSPN KLRC1 PTMS SCML4 ZBTB10 CCL3 CA5B PIP5K1B WNT9A CCNH GEM IL18RAP GGH SARDH B3GNT7 C13orf46 SBF2 IKZF3 ZMAT1 TCF7 NECTIN1 H3C7 FOS PAG1 HECA SLC4A10 SLC35G2 PER1 P2RY1 NFKBIA WDR76 PLAUR KDM1A H1-5 TSHZ2 FAM102B HMMR GPR132 CCRL2 PARP8 A2M ST8SIA1 NUF2 IL5RA RBPMS UBE2T USP53 EEF1A1 PLAC8 LGR6 TMEM123 NEK2 SNAP47 PTGIS SH2B3 P2RY8 S100PBP PLEKHA7 CLNK CRIM1 MGAT5 YBX3 TP53INP1 DTL CFH FEZ1 MYB FRMD4B TSPAN5 STIL ITGA2 GOLGA6L10 MYBL2 AHI1 CAND2 GZMB RBPJ PELI1 HSPA1B KCNK5 GOLGA6L9 TICRR TPRG1 UBE2C AURKA Leem G, et al.