Tensorflow 2.0 and Kubeflow for Scalable and Reproducable Enterprise Ai
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Running ML/DL Workloads Using Red Hat Openshift Container Platform V3.11 Accelerate Your ML/DL Projects Platform Using Kubeflow and NVIDIA Gpus
Running ML/DL Workloads Using Red Hat OpenShift Container Platform v3.11 Accelerate your ML/DL Projects Platform using Kubeflow and NVIDIA GPUs January 2021 H17913.2 White Paper Abstract This white paper describes how to deploy Kubeflow v0.5 on Red Hat OpenShift Container Platform and provides recommendations for achieving optimal performance for ML/DL workloads using the latest NVIDIA Tesla GPUs. Dell Technologies Solutions Copyright The information in this publication is provided as is. Dell Inc. makes no representations or warranties of any kind with respect to the information in this publication, and specifically disclaims implied warranties of merchantability or fitness for a particular purpose. Use, copying, and distribution of any software described in this publication requires an applicable software license. Copyright © 2021 Dell Inc. or its subsidiaries. All Rights Reserved. Dell Technologies, Dell, EMC, Dell EMC and other trademarks are trademarks of Dell Inc. or its subsidiaries. Intel, the Intel logo, the Intel Inside logo and Xeon are trademarks of Intel Corporation in the U.S. and/or other countries. Other trademarks may be trademarks of their respective owners. Published in the USA 01/21 White Paper H17913.2. Dell Inc. believes the information in this document is accurate as of its publication date. The information is subject to change without notice. 2 Running ML/DL Workloads Using Red Hat OpenShift Container Platform v3.11 Accelerate your ML/DL Projects Platform using Kubeflow and NVIDIA GPUs White Paper Contents Contents Executive summary ........................................................................................................................ 4 Solution architecture ...................................................................................................................... 6 GPU accelerated TensorFlow training using TFJobs .................................................................. 9 Installing the GPU device plug-in ............................................................................................... -

Application Development with Azure
Application Development with Azure Karim Vaes Specialist – Azure Application Development 0032 497 219577 @kvaes Agenda • Kubernetes Kubernetes Kubernetes momentum “By 2020, more than 50% of enterprises Larger companies will run mission-critical, containerized are leading the cloud-native applications in production.” adoption. 77% For the organizations running Kubernetes today, 77%1 of those with more than 1,000 developers are running it in production. 1Heptio: state of Kubernetes 2018 What’s behind the growth? Kubernetes: the leading orchestrator shaping the future app development and management It’s widely used It’s vendor-neutral It’s community-supported Kubernetes is in production for A variety of cloud providers There’s a huge community of active global companies across industries1 offer robust Kubernetes support contributors supporting Kubernetes3 24,000 1.1 million contributors contributions since 2016 since 2016 1Kubernetes.io. “Kubernetes User Case Studies.” 2CNCF. “Kubernetes Is First…” 3CNCF. Keynote address. Azure Kubernetes Service (AKS) Ship faster, operate easily, and scale confidently with managed Kubernetes on Azure Manage Kubernetes Accelerate Build on an Run anything, with ease containerized enterprise-grade, anywhere development secure foundation Top scenarios for Kubernetes on Azure Lift and shift Machine Microservices IoT Secure DevOps to containers learning Cost saving Agility Performance Portability Automation without refactoring Faster application Low latency Build once, Deliver code faster and your app development processing run anywhere securely at scale Azure Kubernetes momentum Trusted by thousands of customers 30x Azure Kubernetes Service usage grew 30x since it was made generally available in June 2018 Dated November 2018 How Kubernetes works Kubernetes control Worker node Internet kubelet kube-proxy 1. -

Running Large-Scale Machine Learning Experiments in the Cloud
O’REILLY Artificial Intelligence Conference, San Jose 2019 Running large-scale machine learning experiments in the cloud Shashank Prasanna, Amazon Web Services @shshnkp September 12, 2019 © 2019, Amazon Web Services, Inc. or its Affiliates. All rights reserved. Agenda • Machine Learning, experiments and the scientific method • Scaling challenges with machine learning setups • Containers technologies for large-scale machine learning • Demo 1: Hyperparameter optimization with Amazon SageMaker • Demo 2: Custom designed experiments with Amazon SageMaker • Demo 3: Hyperparameter optimization with Kubernetes, Kubeflow and Katib • Summary and additional resources © 2019, Amazon Web Services, Inc. or its Affiliates. All rights reserved. Experimentation in machine learning Data acquisition Data preparation for Design and run curation and labeling training experiments Model optimization Distributed Deployment and validation training © 2019, Amazon Web Services, Inc. or its Affiliates. All rights reserved. Why do we run experiments? The scientific method Question Hypothesis empirical procedure to determine Experiment whether observations agree or conflict with our hypothesis Interpret Iterate © 2019, Amazon Web Services, Inc. or its Affiliates. All rights reserved. Why do we run machine learning experiments? Machine learning researcher may want to: Question • Identify factors that affect model performance Hypothesis • Explain the root causes of performance Experiment • Choose between alternate models Interpret • Study variability and robustness of -

Chapter 1 - Overview
Reference architecture Chapter 1 - Overview This document provides a complete reference architecture guide for infrastructure required for machine learning use cases. This document specifically covers open source machine learning software that includes Kubeflow using Charmed Kubernetes solution on Dell EMC hardware delivered by Canonical. This includes Dell EMC PowerEdge servers for workloads and storage based on Intel® Xeon® Scalable Processors and Dell EMC Networking. This guide discusses the Dell EMC hardware specifications and the tools and services to set up both the hardware and software, including the foundation cluster and the Kubernetes cluster to run machine learning workloads. It also covers other tools used for the monitoring and management of the cluster in detail and how all these components work together in the system. The guide also provides the deployment steps and references to configuration and automation scripts developed by Dell EMC and Canonical for the deployment process. Document ID Revisions Revisions Date Description March 2020 Initial release Acknowledgements This paper was produced by the following: Author: Andrey Grebennikov Support: Other: The information in this publication is provided “as is.” Dell Inc. makes no representations or warranties of any kind with respect to the information in this publication, and specifically disclaims implied warranties of merchantability or fitness for a particular purpose. Use, copying, and distribution of any software described in this publication requires an applicable software license. Copyright © Dell Inc. or its subsidiaries. All Rights Reserved. Dell, EMC, Dell EMC and other trademarks are trademarks of Dell Inc. or its subsidiaries. Other trademarks may be trademarks of their respective owners. -

Using Ipus from Docker Release 1.4.0
Using IPUs from Docker Release 1.4.0 Graphcore Ltd Dec 08, 2020 CONTENTS 1 Introduction 1 2 Initial setup 2 3 Using gc-docker 3 3.1 Loading docker images.............................................3 3.2 Verifying IPU access from inside container..................................4 3.3 Mounting directories from the host......................................5 3.4 Setting environment variables.........................................5 4 Running a TensorFlow application on an IPU6 5 Extending the images 7 6 Further reading 8 7 Trademarks & copyright 9 i CHAPTER ONE INTRODUCTION This guide explains how you can run applications in Docker on a Linux machine with one or more physical IPU devices. Prerequisites: • A machine with IPU devices • Ubuntu 18.04 / CentOS 7.6 1 CHAPTER TWO INITIAL SETUP First check if your machine has the IPU device driver installed. You can check this is loaded and running with the following command: $ modinfo ipu_driver If the driver is are installed and running, you should see something similar to: $ modinfo ipu_driver filename: /lib/modules/4.15.0-55-generic/updates/dkms/ipu_driver.ko version: 1.0.39 description: IPU PCI Driver author: Graphcore Limited license: GPL srcversion: 49FFB7D8556EB58899AE41A alias: pci:v00001D95d00000003sv*sd*bc*sc*i* alias: pci:v00001D95d00000002sv*sd*bc*sc*i* alias: pci:v00001D95d00000001sv*sd*bc*sc*i* depends: retpoline: Y name: ipu_driver vermagic: 4.15.0-55-generic SMP mod_unload parm: memmap_start:array of ulong parm: memmap_size:array of ulong If so, proceed to the next section. If it returns an error along the lines of: $ modinfo ipu_driver modinfo: ERROR: Module ipu_driver not found. You will need to install the driver. -

Reference Architecture for Kubeflow on Openshift Accelerate ML/DL Workloads Using Kubeflow and Poweredge Servers
Solution brief Reference Architecture for Kubeflow on OpenShift Accelerate ML/DL workloads using Kubeflow and PowerEdge servers Artificial intelligence (AI) and its subsets, machine learning (ML) and deep learning (DL), Customer are gaining widespread adoption for use cases such as computer vision, speech recognition and natural language processing (NLP). Integrating production‑grade AI technologies Results in well‑defined platforms within the protection of the data center can facilitate wider adoption of advanced computing, extending investments by supporting AI use cases as well as augmenting the resources available to data science teams. 2 hours vs. But not every IT organization has the time and resources to research, integrate and test all the components required to deploy a customized system to run AI workloads. Building 9 months a production‑ready AI system involves combining various components, often from different to run analysis1 vendors, and integrating and managing these disparate pieces. As well, deployments are often tied to a specific cluster, so that moving models — for example, between laptops and cloud clusters or from DevOps to production and back — significantly increases 218% ROI complexity and the chance for human errors. over 3 years2 Kubeflow together with the Red Hat® OpenShift® Container Platform help address these challenges. Kubeflow is an open‑source Kubernetes®‑native platform designed 20 million to accelerate ML workloads. It’s a composable, scalable, portable stack that includes images used to train a components and automation features to integrate ML tools, so they work together to deep neural network3 create a cohesive pipeline that makes it easy to deploy ML applications at scale. -

CNCF Webinar Taming Your AI/ML Workloads with Kubeflow
CNCF Webinar Taming Your AI/ML Workloads with Kubeflow The Journey to Version 1.0 David Aronchick (Microsoft) - @aronchick Elvira Dzhuraeva (Cisco) - @elvirafortune Johnu George (Cisco) - @johnugeorge SNARC Maze Solver Minsky / Edmonds (1951) 2000 3 2006 4 2007 5 2008 6 2010 7 2013 8 2014 9 2015 10 Today 11 One More ML ???Solution??? 12 One More ML ???Solution??? 13 14 GitHub Natural Language Search Prototype MVP With Demo In Jupyter Demo with front-end mockup with Experiments.Github.Com: +3 Months Notebook: 2 Weeks blog post: +3 Days https://github.com/hamelsmu/code_search https://towardsdatascience.com/semantic-code-se https://experiments.github.com/ arch-3cd6d244a39c 15 Resource Provisioning Numpy Scheduling Experiment Tracking Jupyter Access Control Declarative Deployment TF.Transform Drivers HP Tuning TF.Estimator Orchestration Profiling Docker Lifecycle Management Validation Seldon Networking KubeCon 2017 17 Resource Provisioning Numpy Scheduling Experiment Tracking Jupyter Access Control Declarative Deployment TF.Transform Drivers HP Tuning TF.Estimator Orchestration Profiling Docker Lifecycle Management Validation Seldon Networking Mission (2017) Make it Easy for Everyone to Develop, Deploy and Manage Portable, Distributed ML on Kubernetes 20 Kubeflow 2018 Mission (2018) Make it Easy for Everyone to Develop, Deploy and Manage Portable, Distributed ML on Kubernetes 23 Kubeflow 2019 Mission (2019) Make it Easy for Everyone to Develop, Deploy and Manage Portable, Distributed ML on Kubernetes 26 Mission (2020) Make it Easy for Everyone -
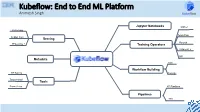
Kubeflow: End to End ML Platform Animesh Singh
Kubeflow: End to End ML Platform Animesh Singh Jupyter Notebooks MXNet KFServing Tensorflow Seldon Core Serving Pytorch TFServing, + Training Operators XGBoost, + MPI Metadata Kale Workflow Building HP Tuning Fairing Tensorboard Tools Prometheus KF Pipelines Pipelines TFX Your Speaker Today: CODAIT 2 Animesh Singh Kubeflow STSM and Chief Architect - Data and AI Open Source github.com/kubeflow Platform o CTO, IBM RedHat Data and AI Open Source Alignment o IBM Kubeflow Engagement Lead, Kubeflow Committer o Chair, Linux Foundation AI - Trusted AI o Chair, CD Foundation MLOps Sig o Ambassador, CNCF o Member of IBM Academy of Technology (IBM AoT) © 2019 IBM Corporation Kubeflow: Current IBM Contributors Untrained Model Christian Kadner Weiqiang Zhuang Tommy Li Andrew Butler Prepared Preparedand Trained AnalyzedData Model Data Deployed Model Jin Chi He Feng Li Ke Zhu Kevin Yu IBM is the 2nd Largest Contributor IBM is the 2nd Largest Contributor IBMers contributing across projects in Kubeflow Kubeflow Services kubectl apply -f tfjob High Level Kubernetes API Server Services Notebooks Low Level APIs / Services Pipelines TFJob PyTorchJob Pipelines CR KFServing Argo Jupyter CR MPIJob Katib Seldon CR Study Job Spark Job Istio Mesh and Gateway Kubebench TFX Developed By Kubeflow Developed Outside Kubeflow Adapted from Kubeflow Contributor Summit 2019 talk: Kubeflow and ML Landscape (Not all components are shown) Community is growing! 8 Multi-User Isolation ML Lifecycle: Build: Development, Training and HPO Untrained Model Prepared Preparedand Trained -

Machine Learning at Scale with Kubernetes Aug 23Rd, 2018
JupyterCon - Machine Learning at Scale with Kubernetes Aug 23rd, 2018 Chris Cho, Fei Xue, Tyler Erickson Confidential & Proprietary Proprietary + Confidential Contents 1. Containers Appendix- Tensor2Tensor 2. Kubernetes 3. Google Kubernetes Engine 4. Kubeflow - Overview 5. Kubeflow - Components 6. Kubeflow - Architecture 7. ksonnet 8. Kubeflow - Tutorial Section Start Time Total Duration (Min) Type Topic area Proprietary + Confidential Agenda 9:00 AM 0:30 Presentation Welcome + Introduction + Setup 9:30 AM 0:45 Exercise GKE quest lab 1- Introduction to docker 10:15 AM 0:05 Debrief 10:20 AM 0:30 Presentation Kubernetes and GKE 10:50 AM 1:00 Exercise GKE quest lab 2 - Hello Node Kubernetes 11:50 AM 0:05 Debrief 11:55 AM 0:30 Presentation Orchestrating the Cloud with GKE 12:25 PM 5:25 1:00 Lunch 1:25 PM 1:00 Exercise GKE quest lab 3 - Orchestrating the Cloud with GKE 1:25 PM 0:30 Presentation Kubeflow introduction 1:55 PM 1:00 Exercise Kubeflow 1 - Intro to Kubeflow 2:55 PM 0:05 Debrief 3:00 PM 0:30 Presentation Kubeflow - Components, Architecture, Short tutorial 3:00 PM 1:30 Exercise Kubeflow 2 - E2E 4:30 PM 3:35 Buffer Proprietary + Confidential Expectations – Walk away with better understanding of… ● Containers/Docker ● Kubernetes ● Orchestrating Kubernetes ● Kubeflow – Try out Kubeflow when you get home :) Proprietary + Confidential Logistics – Access you will need for the labs ● 8 Qwiklab credits - Pick it up in the back ● 2 GCP test accounts - Handed out at the end – Sign up on https://qwiklabs.com – Enroll in quest https://qwiklabs.com/quests/29 -

Application Development with Azure
Application Development with Azure Karim Vaes Specialist – Azure Application Development @kvaes Agenda • Digital Transformation, powered by Application Innovation • Developer Toolchain • App Service • Integration Services Digital Transformation Powered by Application Innovation Digital transformation 91% Digital of business leaders see Digital Transformation as a way of sparking Transformation innovation and finding efficiencies1 A journey with one destination but different paths 85% say they must offer digital services or become irrelevant2 1 ISACA: Information Systems Audit and Control Association, 2018 2 Couchbase: Couchbase Survey, August 2018 1 Data: Capture digital signal from across business Consumer Reports review indicate braking issue with Model 3 Vehicle telemetry shows brake performance across fleet 2 Insight: Connect and synthesize data Car telemetry for suspect cars analyzed to understand issue Tesla identifies fix to improve stopping distance Engage Transform customers products 3 Action: Improve business outcomes Car braking software updated over-the-air to fix issue Tesla closes the loop with consumer reports and review is updated 7,0% 6,0% 5,0% 4,0% 3,0% 2,0% 1,0% 0,0% -1,0% -2,0% software Digital DNA Toolchain Overview World’s most comprehensive developer toolchain Azure Azure Stack Azure Data Box Azure Sphere Azure Kinect HoloLens Web Databases Mobile Analytics Tools Mixed Reality AI + Machine Learning Visual Studio Containers Internet of Things Azure Devops Events + Integration Media GitHub PowerApps Power BI Compute -

Build an Event Driven Machine Learning Pipeline on Kubernetes
Assign Hyperparameters Initial Model and Train Create Model PreparedPrepared andand Trained AnalyzedAnalyzed Model DataData Monitor DeployedDeployed Validate and Deploy ModelModel Build an Event Driven Machine Learning Pipeline on Kubernetes Yasushi Osonoi Animesh Singh Developer Advocate IBM STSM, IBM kubeflow kfserving maintainer osonoi animeshsingh Center for Open Source Improving Enterprise AI lifecycle in Open Source Data and AI Technologies (CODAIT) Code – Build and improve practical frameworks to enable more developers to realize immediate value. Content – Showcase solutions for complex and real-world AI problems. Community – Bring developers and data scientists to engage with IBM • Team contributes to over 10 open source projects • 17 committers and many contributors in Apache projects • Over 1100 JIRAs and 66,000 lines of code committed to Apache Spark itself; over 65,000 LoC into SystemML • Over 25 product lines within IBM leveraging Apache Spark • Speakers at over 100 conferences, meetups, unconferences and more CODAIT codait.org 3 DEVELOPER ADVOCATE in TOKYO Tokyo Team is a part of Worldwide Developer Advocate Teams! Developer Advocate City Leader WW Developer Advocate WW Developer Advocate Client Developer Advocate AKIRA ONISHI NORIKO KATO KYOKO NISHITO YASUSHI OSONOI Program Manager WW Developer Advocate WW Developer Advocate Digital Developer Advocate TOSHIO YAMASHITA TAIJI HAGINO AYA TOKURA JUNKI SAGAWA @taiponrock https://developer.ibm.com/patterns/ https://developer.ibm.com/jp/ Please follow me @osonoi IBM’s history -
![Arxiv:2103.00490V1 [Cs.DC] 24 Feb 2021 Keywords: Dataset Lifecycle Framework · Kubeflow · Kubernetes · Bioinformatics](https://docslib.b-cdn.net/cover/8795/arxiv-2103-00490v1-cs-dc-24-feb-2021-keywords-dataset-lifecycle-framework-%C2%B7-kube-ow-%C2%B7-kubernetes-%C2%B7-bioinformatics-2588795.webp)
Arxiv:2103.00490V1 [Cs.DC] 24 Feb 2021 Keywords: Dataset Lifecycle Framework · Kubeflow · Kubernetes · Bioinformatics
Dataset Lifecycle Framework and its applications in Bioinformatics Yiannis Gkoufas1 and David Yu Yuan, Ph.D.2[0000−0003−1075−1628] 1 IBM Research - Ireland [email protected] https://www.ibm.com/ 2 Technology and Science Integration, European Bioinformatics Institute, European Molecular Biology Laboratory [email protected] https://www.ebi.ac.uk/ Abstract. Bioinformatics pipelines depend on shared POSIX filesys- tems for its input, output and intermediate data storage. Containeriza- tion makes it more difficult for the workloads to access the shared file systems. In our previous study, we were able to run both ML and non- ML pipelines on Kubeflow successfully. However, the storage solutions were complex and less optimal. This is because there are no established resource types to represent the concept of data source on Kubernetes. More and more applications are running on Kubernetes for batch pro- cessing. End users are burdened with configuring and optimising the data access, which is what we have experienced before. In this article, we are introducing a new concept of Dataset and its corresponding resource as a native Kubernetes object. We have leveraged the Dataset Lifecycle Framework which takes care of all the low-level details about data access in Kubernetes pods. Its pluggable architecture is designed for the development of caching, scheduling and governance plugins. Together, they manage the entire lifecycle of the custom resource Dataset. We use Dataset Lifecycle Framework to serve data from object stores to both ML and non-ML pipelines running on Kubeflow. With DLF, we make training data fed into ML models directly without being down- loaded to the local disks, which makes the input scalable.