Emodnet Phase 2 – Final Report Reporting Period: 01/09/2013 – 01/09/2016
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

THE DEVELOPMENT of MESOZOIC SEDIMENTARY IBASINS AROUND the MARGINS of the NORTH ATLANTIC and THEIR HYDROCARBON POTENTIAL D.G. Masson and P.R
INTERNAL DOCUMENT IIL THE DEVELOPMENT OF MESOZOIC SEDIMENTARY IBASINS AROUND THE MARGINS OF THE NORTH ATLANTIC AND THEIR HYDROCARBON POTENTIAL D.G. Masson and P.R. Miles Internal Document No. 195 December 1983 [This document should not be cited in a published bibliography, and is supplied for the use of the recipient only]. iimsTit\jte of \ OCEANOGRAPHIC SCIENCES % INSTITUTE OF OCEANOGRAPHIC SCIENCES Wormley, Godalming, Surrey GU8 5UB (042-879-4141) (Director: Dr. A. S. Laughton, FRS) Bidston Observatory, Crossway, Birkenhead, Taunton, Mersey side L43 7RA Somerset TA1 2DW (051-653-8633) (0823-86211) (Assistant Director: Dr. D. E. Cartwright) (Assistant Director: M.J. Tucker) THE DEVELOPMENT OF MESOZOIC SEDIMENTARY BASINS AROUND THE MARGINS OF THE NORTH ATLANTIC AND THEIR HYDROCARBON POTENTIAL D.G. Masson and P.R. Miles Internal Document No. 195 December 1983 Work carried out under contract to the Department of Energy This document should not be cited in a published bibliography except as 'personal communication', and is supplied for the use of the recipient only. Institute of Oceanographic Sciences, Brook Road, Wormley, Surrey GU8 SUB ABSTRACT The distribution of Mesozoic basins around the margins of the North Atlantic has been summarised, and is discussed in terms of a new earliest Cretaceous palaeogeographic reconstruction for the North Atlantic area. The Late Triassic-Early Jurassic rift basins of Iberia, offshore eastern Canada and the continental shelf of western Europe are seen to be the fragments of a formerly coherent NE trending rift system which probably formed as a result of tensional stress between Europe, Africa and North America. The separation of Europe, North America and Iberia was preceded by a Late Jurassic-Early Cretaceous rifting phase which is clearly distinct from the earlier Mesozoic rifting episode and was little influenced by it. -
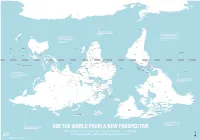
Is Map Is Just As Accurate As the One We're All Used To
ROSS SEA WEDDELL SEA AMUNDSEN SEA ANTARCTICA BELLINGSHAUSEN SEA AMERY ICE SHELF SOUTHERN OCEAN SOUTHERN OCEAN SOUTHERN OCEAN SCOTIA SEA DRAKE PASSAGE FALKLAND ISLANDS Stanley (U.K.) THE RATIO OF LAND TO WATER IN THE SOUTHERN HEMISPHERE BY THE TIME EUROPEANS ADOPTED IS 1 TO 5 THE NORTH-POINTING COMPASS, PTOLEMY WAS A HELLENIC Wellington THEY WERE ALREADY EXPERIENCED NEW TASMAN SEA ZEALAND CARTOGRAPHER WHOSE WORK CHILE IN NAVIGATING WITH REFERENCE TO IN THE SECOND CENTURY A.D. ARGENTINA THE NORTH STAR Canberra Buenos Santiago GREAT AUSTRALIAN BIGHT POPULARIZED NORTH-UP Montevideo Aires SOUTH PACIFIC OCEAN SOUTH ATLANTIC OCEAN URUGUAY ORIENTATION SOUTH AFRICA Maseru LESOTHO SWAZILAND Mbabane Maputo Asunción AUSTRALIA Pretoria Gaborone NEW Windhoek PARAGUAY TONGA Nouméa CALEDONIA BOTSWANA Saint Denis NAMIBIA Nuku’Alofa (FRANCE) MAURITIUS Port Louis Antananarivo MOZAMBIQUE CHANNEL ZIMBABWE Suva Port Vila MADAGASCAR VANUATU MOZAMBIQUE Harare La Paz INDIAN OCEAN Brasília LAKE SOUTH PACIFIC OCEAN FIJI Lusaka TITICACA CORAL SEA GREAT GULF OF BARRIER CARPENTARIA Lilongwe ZAMBIA BOLIVIA FRENCH POLYNESIA Apia REEF LAKE (FRANCE) SAMOA TIMOR SEA COMOROS NYASA ANGOLA Lima Moroni MALAWI Honiara ARAFURA SEA BRAZIL TIMOR LESTE PERU Funafuti SOLOMON Port Dili Luanda ISLANDS Moresby LAKE Dodoma TANGANYIKA TUVALU PAPUA Jakarta SEYCHELLES TANZANIA NEW GUINEA Kinshasa Victoria BURUNDI Bujumbura DEMOCRATIC KIRIBATI Brazzaville Kigali REPUBLIC LAKE OF THE CONGO SÃO TOMÉ Nairobi RWANDA GABON ECUADOR EQUATOR INDONESIA VICTORIA REP. OF AND PRINCIPE KIRIBATI EQUATOR -

Chapter 24. the BAY of BISCAY: the ENCOUNTERING of the OCEAN and the SHELF (18B,E)
Chapter 24. THE BAY OF BISCAY: THE ENCOUNTERING OF THE OCEAN AND THE SHELF (18b,E) ALICIA LAVIN, LUIS VALDES, FRANCISCO SANCHEZ, PABLO ABAUNZA Instituto Español de Oceanografía (IEO) ANDRE FOREST, JEAN BOUCHER, PASCAL LAZURE, ANNE-MARIE JEGOU Institut Français de Recherche pour l’Exploitation de la MER (IFREMER) Contents 1. Introduction 2. Geography of the Bay of Biscay 3. Hydrography 4. Biology of the Pelagic Ecosystem 5. Biology of Fishes and Main Fisheries 6. Changes and risks to the Bay of Biscay Marine Ecosystem 7. Concluding remarks Bibliography 1. Introduction The Bay of Biscay is an arm of the Atlantic Ocean, indenting the coast of W Europe from NW France (Offshore of Brittany) to NW Spain (Galicia). Tradition- ally the southern limit is considered to be Cape Ortegal in NW Spain, but in this contribution we follow the criterion of other authors (i.e. Sánchez and Olaso, 2004) that extends the southern limit up to Cape Finisterre, at 43∞ N latitude, in order to get a more consistent analysis of oceanographic, geomorphological and biological characteristics observed in the bay. The Bay of Biscay forms a fairly regular curve, broken on the French coast by the estuaries of the rivers (i.e. Loire and Gironde). The southeastern shore is straight and sandy whereas the Spanish coast is rugged and its northwest part is characterized by many large V-shaped coastal inlets (rias) (Evans and Prego, 2003). The area has been identified as a unit since Roman times, when it was called Sinus Aquitanicus, Sinus Cantabricus or Cantaber Oceanus. The coast has been inhabited since prehistoric times and nowadays the region supports an important population (Valdés and Lavín, 2002) with various noteworthy commercial and fishing ports (i.e. -

Names of Sub-Areas and Divisions of FAO Fishing Areas 27 and 37 NORTH-EAST ATLANTIC
Names of Sub-areas and Divisions of FAO fishing areas 27 and 37 NORTH-EAST ATLANTIC Subarea I Barents Sea Subarea II Norwegian Sea, Spitzbergen, and Bear Island Division II a Norwegian Sea Division II b Spitzbergen and Bear Island Subarea III Skagerrak, Kattegat, Sound, Belt Sea, and Baltic Sea; the Sound and Belt together known also as the Transition Area Division III a Skagerrak and Kattegat Division III b,c Sound and Belt Sea or Transition Area Division III b (23) Sound Division III c (22) Belt Sea Division III d (24-32) Baltic Sea Subarea IV North Sea Division IV a Northern North Sea Division IV b Central North Sea Division IV c Southern North Sea Subarea V Iceland and Faroes Grounds Division V a Iceland Grounds Division V b Faroes Grounds Subarea VI Rockall, Northwest Coast of Scotland and North Ireland, the Northwest Coast of Scotland and North Ireland also known as the West of Scotland Division VI a Northwest Coast of Scotland and North Ireland or West of Scotland Division VI b Rockall Subarea VII Irish Sea, West of Ireland, Porcupine Bank, Eastern and Western English Channel, Bristol Channel, Celtic Sea North and South, and Southwest of Ireland - East and West Division VII a Irish Sea Division VII b West of Ireland Division VII c Porcupine Bank Division VII d Eastern English Channel Division VII e Western English Channel Division VII f Bristol Channel Division VII g Celtic Sea North Division VII h Celtic Sea South Division VII j South-West of Ireland - East Division VII k South-West of Ireland - West Subarea VIII Bay of Biscay -

Pathways of Education for Gender Equality
Pathways of Education for Gender Equality Research Report on the project Gender Equality, Unemployment and Lifelong Learning in Europe – Impact on Life Chances of Young Women Zagreb, 2015 Impressum Publisher: Centre for Women’s Studies, Dolac 8, Zagreb Phone: +385 1 4872-406; www.zenstud.hr For the publisher: Rada Borić Editing: Sandra Prlenda and Leda Sutlović Layout: Lejla Topić Cover: Susan Jakopec Printed copies: 100 Printed by: Print Studio d.o.o. ISBN: 978-953-6955-51-0 Zagreb, July 2015 Authors: Casa internazionale delle donne, Trieste, Italy Melita Richter, Mira Oklobdžija and Stefania Marchese, authors Centre for Women’s Studies, Zagreb, Croatia Leda Sutlović and Zorica Siročić, authors Stephanie Olson, proofreading Mediterranean Institute for Gender Studies, Nicosia, Cyprus Susana Pavlou and Christina Demetriades, authors This research was created within the “Gender Equality, Unemployment and Lifelong Learning in Europe – Impact on Life Chances of Young Women”, Grundtvig Learning Partnership Project. The information and views set out in this publication are those of the authors and do not necessarily reflect the official opinion of the European Union. Table of Contents /Preface .................................................................................................................................................. 4 /Leda Sutlović and Zorica Siročić Unfinished Institutionalisation – Women’s and Gender Studies in Croatia – research report .................................................................................................................................. -

Temporal Patterns in Habitat Use by Small Cetaceans at an Oceanographically Dynamic Marine Renewable Energy Test Site in the Celtic Sea
Deep-Sea Research II ∎ (∎∎∎∎) ∎∎∎–∎∎∎ Contents lists available at ScienceDirect Deep-Sea Research II journal homepage: www.elsevier.com/locate/dsr2 SI: European Marine Megafauna Temporal patterns in habitat use by small cetaceans at an oceanographically dynamic marine renewable energy test site in the Celtic Sea S.L. Cox a,b,n, M.J. Witt c, C.B. Embling a,d, B.J. Godley d, P.J. Hosegood b, P.I. Miller e, S.C. Votier c, S.N. Ingram a a Marine Biology and Ecology Research Centre, Plymouth University, Plymouth PL4 8AA, UK b Marine Physics Research Group, Plymouth University, Plymouth PL4 8AA, UK c Environment and Sustainability Institute, University of Exeter, Penryn TR10 9FE, UK d Centre for Ecology and Conservation, College of Life Sciences, University of Exeter, Penryn TR10 9FE, UK e Plymouth Marine Laboratory, Prospect Place, Plymouth PL1 3DH, UK article info abstract Shelf-seas are highly dynamic and oceanographically complex environments, which likely influences the Keywords: spatio-temporal distributions of marine megafauna such as marine mammals. As such, understanding Marine predator natural patterns in habitat use by these animals is essential when attempting to ascertain and assess the Habitat use impacts of anthropogenically induced disturbances, such as those associated with marine renewable Fronts energy installations (MREIs). This study uses a five year (2009–2013) passive acoustics (C-POD) dataset to Passive acoustics examine the use of an oceanographically dynamic marine renewable energy test site by small cetaceans, Marine -

Annual Variation in Diets, Feeding Locations and Foraging Behaviour of Gannets in the North Sea: Flexibility, Consistency and Constraint
MARINE ECOLOGY PROGRESS SERIES Vol. 338: 295–305, 2007 Published May 24 Mar Ecol Prog Ser Annual variation in diets, feeding locations and foraging behaviour of gannets in the North Sea: flexibility, consistency and constraint K. C. Hamer1,*, E. M. Humphreys1, 2, S. Garthe3, J. Hennicke4, G. Peters5, D. Grémillet6, R. A. Phillips7, M. P. Harris8, S. Wanless8 1Earth Biosphere Institute and Faculty of Biological Sciences, University of Leeds, Leeds LS2 9JT, UK 2British Trust for Ornithology Scotland, School of Biological and Environmental Sciences, Stirling University, Stirling FK9 4LA, UK 3Centre for Research and Technology Westkuste, University of Kiel, 25761 Büsum, Germany 4Institute of Zoology, University of Hamburg, 20146 Hamburg, Germany 5Earth & Ocean Technologies, Krummbogen 32, 24113 Kiel, Germany 6Percy FitzPatrick Institute, University of Cape Town, Rondebosch 7701, South Africa 7NERC British Antarctic Survey, Madingley Road, Cambridge CB3 0ET, UK 8NERC Centre for Ecology and Hydrology, Banchory Research Station, Hill of Brathens, Aberdeenshire AB31 4BY, UK ABSTRACT: Many seabirds nesting in areas bordering the North Sea have recently experienced large annual variation in breeding success, including reproductive failures in some cases. In contrast, the breeding success of northern gannets Morus bassanus has remained remarkably stable. The pre- sent study examines data from the large gannet colony at the Bass Rock (southeast Scotland) across 3 years, to assess the extent to which such stability may reflect both flexibility and consistency in diets and foraging behaviour. Adults exhibited great flexibility both in the species and sizes of prey con- sumed and in foraging trip durations, ranges and total distances travelled. They also showed a high degree of consistency in bearings of foraging trips and in behaviour at sea; the sinuosity of foraging tracks and average speed of travel was very similar each year and birds in all years spent about half their time at sea in flight. -

Università Degli Studi Di Trieste Xxx Ciclo Del
UNIVERSITÀ DEGLI STUDI DI TRIESTE XXX CICLO DEL DOTTORATO DI RICERCA IN Scienze della Riproduzione e dello Sviluppo IDENTIFICATION OF GENETIC VARIANTS REGULATING FEMALE FERTILITY Settore scientifico-disciplinare: MED/03 DOTTORANDA: Caterina Maria Barbieri COORDINATORE: PROF. Paolo Gasparini SUPERVISORE DI TESI: PROF.SSA Daniela Toniolo PROF. Paolo Gasparini ANNO ACCADEMICO 2016/2017 UNIVERSITÀ DEGLI STUDI DI TRIESTE XXX CICLO DEL DOTTORATO DI RICERCA IN Scienze della Riproduzione e dello Sviluppo IDENTIFICATION OF GENETIC VARIANTS REGULATING FEMALE FERTILITY Settore scientifico-disciplinare: MED/03 DOTTORANDA: Caterina Maria Barbieri COORDINATORE: PROF. Paolo Gasparini SUPERVISORE DI TESI: PROF.SSA Daniela Toniolo PROF. Paolo Gasparini ANNO ACCADEMICO 2016/201 CONTENTS CONTENTS ............................................................................................................................ 4 LIST OF ABBREVIATION .................................................................................................. 6 INTRODUCTION .................................................................................................................. 7 Women fertility ........................................................................................................................... 8 Follicle development and ovarian reserve ...................................................................... 10 Age-related decline of female fertility .............................................................................. 12 Anti Mϋllerian -

Atlantic Herring (Clupea Harengus) in the Irish and Celtic Seas; Tracing Populations of the Past and Present
Atlantic herring (Clupea harengus) in the Irish and Celtic Seas; tracing populations of the past and present Nóirín D. Burke A thesis submitted to the School of Science, Galway Mayo Institute of Technology in fulfilment for the degree of Doctor of Philosophy Department of Life and Physical Sciences June 2008 Head of Department: Dr Seamus Lennon Supervisors: Dr Deirdre Brophy Dr Pauline A. King Galway-Mayo Institute of Technology, Dublin Rd., Galway, Ireland TABLE OF CONTENTS Chapter 1: General Introduction……………………………………………........... 10 1.1 Herring biology and population structure………………………. 10 1.2 Herring fisheries around Ireland………………………………... 12 1.3 Stock structure of Celtic and Irish Sea herring………………….. 14 1.4 Herring assessments in the Irish and Celtic Seas……………….. 15 1.5 Otolith applications in fisheries science………………………… 16 1.6 Otolith microstructure……………...……………………………. 17 1.7 Otolith morphometrics and shape analysis……………………… 18 1.8 Summary of objectives………………………………………….. 19 Chapter 2: Shape analysis of otolith annuli in Atlantic herring (Clupea harengus); a new method for tracking fish populations………………………….. 23 2.1 Abstract………………………………………………………….. 23 2.2 Introduction……………………………………………………… 23 2.3 Methods………………………………………………………….. 26 2.4 Results…………………………………………………………… 32 2.5 Discussion……………………………………………………….. 33 2.6 Conclusion……………………………………………………….. 40 Chapter 3: Otolith shape analysis, its applications to distinguishing between Irish and Celtic Sea herring (Clupea harengus) stocks in the Irish Sea……………….. 49 3.1 Abstract………………………………………………………….. 49 3.2 Introduction……………………………………………………… 49 3.3 Methods…………………………………………………………. 51 3.4 Results…………………………………………………………… 54 3.5 Discussion……………………………………………………….. 55 3.6 Conclusion……………………………………………………….. 57 Chapter 4: Otolith shape analysis provides evidence of natal homing in Atlantic herring (Clupea harengus)………………………………………………………… 62 4.1 Abstract………………………………………………………….. 62 4.2 Introduction……………………………………………………… 62 4.3 Methods…………………………………………………………. -

Celtic Seas Ecoregion – Ecosystem Overview
ICES Ecosystem Overviews Celtic Seas Ecoregion Published 14 December 2018 https://doi.org/ 10.17895/ices.pub.4667 7.1 Celtic Seas Ecoregion – Ecosystem overview Ecoregion description The Celtic Seas ecoregion covers the northwestern shelf seas of the EU. It includes areas of the deeper eastern Atlantic Ocean and coastal seas that are heavily influenced by oceanic inputs. The ecoregion ranges from north of Shetland to Brittany in the south. Three key areas constitute this ecoregion: • the Malin shelf; • the Celtic Sea and west of Ireland; • the Irish Sea. The Celtic Seas ecoregion includes all or parts of the Exclusive Economic Zones (EEZs) of three EU Member States. Fisheries in the Celtic Seas are managed through the EU Common Fisheries Policy (CFP), with fisheries of some stocks managed by the North East Atlantic Fisheries Commission (NEAFC) and by coastal state agreements. Responsibility for salmon fisheries is taken by the North Atlantic Salmon Conservation Organization (NASCO) and for large pelagic fish by the International Commission for the Conservation of Atlantic Tunas (ICCAT). Collective fisheries advice is provided by the International Council for the Exploration of the Sea (ICES), the European Commission’s Scientific Technical and Economic Committee for Fisheries (STECF), and the North Western Waters and Pelagic ACs. Environmental policy is managed by national governments and agencies and by OSPAR; advice is provided by national agencies, OSPAR, the European Environment Agency (EEA), and ICES. International shipping is managed under the International Maritime Organization (IMO). Figure 1 The Celtic Seas ecoregion, showing EEZs, larger offshore Natura 2000 sites, and operational and authorized wind farms. -

Submarine Geomorphology of the Celtic Sea Continental Shelf and The
Submarine geomorphology of the Celtic Sea continental shelf and the southern extent of glaciation on the Atlantic margin of Europe D Praeg, Stephen Mccarron, Dayton Dove, Daniella Accettella, Andrea Cova, Lorenzo Facchin, Xavier Monteys To cite this version: D Praeg, Stephen Mccarron, Dayton Dove, Daniella Accettella, Andrea Cova, et al.. Submarine geomorphology of the Celtic Sea continental shelf and the southern extent of glaciation on the Atlantic margin of Europe. INQUA 2019 - 20th Congress of the International Union for Quaternary Research, Jul 2019, Dublin, Ireland. pp.Abstract P-4004. hal-02363131 HAL Id: hal-02363131 https://hal.archives-ouvertes.fr/hal-02363131 Submitted on 14 Nov 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Submarine geomorphology of the Celtic Sea continental shelf and the southern extent of glaciation on the Atlantic margin of Europe P-4004 Daniel Praeg1,2 ([email protected]), S. McCarron3, Dayton Dove4, Daniella Accettella1, Andrea Cova1, Lorenzo Facchin1, Xavier Monteys5 1 Istituto Nazionale di Oceanografia e di Geofisica Sperimentale (OGS), Trieste, ITALY ; 2 Géoazur (UMR7329 CNRS), Valbonne, FRANCE; 3 Maynooth University, Maynooth, IRELAND; 4 British Geological Survey (BGS), Edinburgh, Scotland, UK; 5 Geological Survey of Ireland (GSI), Dublin, IRELAND Maxima of ice sheets Incisions (>100 m relief) Inset - map of buried or partly infilled Pleistocene ‘incisions’ (>100 m in relief)8 Fig. -

TBS Group: Quotati E Contenti TBS Group: Happy to Be Quoted
TBS Group: quotati e contenti TBS Group: Happy To Be Quoted Reti & Partner | Networks & Partners GLOBALE & LOCALE Global & Local Competitività & Impresa | Competitiveness & Business IL DESIGN AMICO A Friendly Design Ricerca & Tecnologia | Research & Technology TRAPPOLE FOTOVOLTAICHE Photovoltaic Traps Conversando con ... | Talking to ... SABRINA STROLEGO Scienza & Dintorni | Science & Surroundings CAPOLAVORI DA SALVARE Masterpieces to be Preserved Ad ApreA4 23-03-2007 11:42 Pagina 1 C M Y CM MY CY CMY K Colori compositi Editoriale Sincrotrone: Editorial più sicuri con SincRoBot Enzo Moi Synchrotron: nuovo Direttore generale Safer with SincRoBot Enzo Moi, 26 the New Director General 2 Primo Piano Legame Close up d’acciaio Quotati Steel e contenti Alliance Happy 28 To Be Quoted Direttore responsabile: 4 Leo Brattoli Reti & Partner Scovare Comitato editoriale: Networks & Partners gli allergeni alimentari Leo Brattoli, Gabriele Gatti, Globale Detecting Francesca Iannelli, & Locale Food Allergens Gianfranco Paulatto, Stephen Taylor Global 30 Redazione: & Local Consorzio per l’AREA di Ricerca Scientifica e tecnologica di Trieste 8 Troppo belle per il Nobel Padriciano 99 - 34012 tel. 040 375 5221 - 5206 Conversando con… fax 040 226698 Sabrina Strolego European Talking to… Hanno collaborato: Sabrina Strolego Alessandro Deltreppo, Generation Antonella Di Noia, Arianna Grabbio, European 33 Emanuele Karantzoulis, Generation Laura Bibi Palatini, Francesca Petrera, Scienza & Dintorni Roberto Pugliese, Laura Ramacci, 12 Science & Surroundings Simona Regina, Ginevra Tonini, Capolavori Massimo Tosto, Franco Zanini da salvare Erasmus Masterpieces Versione on line: Mundus to be Preserved www.area.trieste.it Erasmus 35 Progetto grafico, Mundus impaginazione e pubblicità: 14 Parkinson Competitività & Impresa e dopamina Competitiveness & Business Aps comunicazione Parkinson’s snc di Aldo Poduie e Federica Zar AREA and Dopamin tel.