PDF Datasheet
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Hypothalamus As a Hub for SARS-Cov-2 Brain Infection and Pathogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis Sreekala Nampoothiri1,2#, Florent Sauve1,2#, Gaëtan Ternier1,2ƒ, Daniela Fernandois1,2 ƒ, Caio Coelho1,2, Monica ImBernon1,2, Eleonora Deligia1,2, Romain PerBet1, Vincent Florent1,2,3, Marc Baroncini1,2, Florence Pasquier1,4, François Trottein5, Claude-Alain Maurage1,2, Virginie Mattot1,2‡, Paolo GiacoBini1,2‡, S. Rasika1,2‡*, Vincent Prevot1,2‡* 1 Univ. Lille, Inserm, CHU Lille, Lille Neuroscience & Cognition, DistAlz, UMR-S 1172, Lille, France 2 LaBoratorY of Development and PlasticitY of the Neuroendocrine Brain, FHU 1000 daYs for health, EGID, School of Medicine, Lille, France 3 Nutrition, Arras General Hospital, Arras, France 4 Centre mémoire ressources et recherche, CHU Lille, LiCEND, Lille, France 5 Univ. Lille, CNRS, INSERM, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for Infection and ImmunitY of Lille (CIIL), Lille, France. # and ƒ These authors contriButed equallY to this work. ‡ These authors directed this work *Correspondence to: [email protected] and [email protected] Short title: Covid-19: the hypothalamic hypothesis 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Olfactory Receptor 3A2/3A3 Rabbit Pab Antibody
Olfactory receptor 3A2/3A3 rabbit pAb antibody Catalog No : Source: Concentration : Mol.Wt. (Da): A18879 Rabbit 1 mg/ml 34275 Applications WB,IF,ELISA Reactivity Human Dilution WB: 1:500 - 1:2000. IF: 1:200 - 1:1000. ELISA: 1:5000. Not yet tested in other applications. Storage -20°C/1 year Specificity Olfactory receptor 3A2/3A3 Polyclonal Antibody detects endogenous levels of Olfactory receptor 3A2/3A3 protein. Source / Purification The antibody was affinity-purified from rabbit antiserum by affinity- chromatography using epitope-specific immunogen. Immunogen The antiserum was produced against synthesized peptide derived from human OR3A2/3. AA range:65-114 Uniprot No P47888/P47893 Alternative names OR3A3; OR3A6; OR3A7; OR3A8P; Olfactory receptor 3A3; Olfactory receptor 17-201; OR17-201; Olfactory receptor 3A6; Olfactory receptor 3A7; Olfactory receptor 3A8; Olfactory receptor OR17-22; OR3A2; OLFRA04; Olfactory receptor 3A2; Olfactory Form Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide. Clonality Polyclonal Isotype IgG Conjugation Background olfactory receptor family 3 subfamily A member 3(OR3A3) Homo sapiens Olfactory receptors interact with odorant molecules in the nose, to initiate a neuronal response that triggers the perception of a smell. The olfactory receptor proteins are members of a large family of G-protein-coupled receptors (GPCR) arising from single coding-exon genes. Olfactory receptors share a 7-transmembrane domain structure with many neurotransmitter and hormone receptors and are responsible for the recognition and G protein- mediated transduction of odorant signals. The olfactory receptor gene family is the largest in the genome. The nomenclature assigned to the olfactory receptor genes and proteins for this organism is independent of other organisms. -

Us 2018 / 0305689 A1
US 20180305689A1 ( 19 ) United States (12 ) Patent Application Publication ( 10) Pub . No. : US 2018 /0305689 A1 Sætrom et al. ( 43 ) Pub . Date: Oct. 25 , 2018 ( 54 ) SARNA COMPOSITIONS AND METHODS OF plication No . 62 /150 , 895 , filed on Apr. 22 , 2015 , USE provisional application No . 62/ 150 ,904 , filed on Apr. 22 , 2015 , provisional application No. 62 / 150 , 908 , (71 ) Applicant: MINA THERAPEUTICS LIMITED , filed on Apr. 22 , 2015 , provisional application No. LONDON (GB ) 62 / 150 , 900 , filed on Apr. 22 , 2015 . (72 ) Inventors : Pål Sætrom , Trondheim (NO ) ; Endre Publication Classification Bakken Stovner , Trondheim (NO ) (51 ) Int . CI. C12N 15 / 113 (2006 .01 ) (21 ) Appl. No. : 15 /568 , 046 (52 ) U . S . CI. (22 ) PCT Filed : Apr. 21 , 2016 CPC .. .. .. C12N 15 / 113 ( 2013 .01 ) ; C12N 2310 / 34 ( 2013. 01 ) ; C12N 2310 /14 (2013 . 01 ) ; C12N ( 86 ) PCT No .: PCT/ GB2016 /051116 2310 / 11 (2013 .01 ) $ 371 ( c ) ( 1 ) , ( 2 ) Date : Oct . 20 , 2017 (57 ) ABSTRACT The invention relates to oligonucleotides , e . g . , saRNAS Related U . S . Application Data useful in upregulating the expression of a target gene and (60 ) Provisional application No . 62 / 150 ,892 , filed on Apr. therapeutic compositions comprising such oligonucleotides . 22 , 2015 , provisional application No . 62 / 150 ,893 , Methods of using the oligonucleotides and the therapeutic filed on Apr. 22 , 2015 , provisional application No . compositions are also provided . 62 / 150 ,897 , filed on Apr. 22 , 2015 , provisional ap Specification includes a Sequence Listing . SARNA sense strand (Fessenger 3 ' SARNA antisense strand (Guide ) Mathew, Si Target antisense RNA transcript, e . g . NAT Target Coding strand Gene Transcription start site ( T55 ) TY{ { ? ? Targeted Target transcript , e . -

OR3A2 Sirna (H): Sc-94106
SANTA CRUZ BIOTECHNOLOGY, INC. OR3A2 siRNA (h): sc-94106 BACKGROUND STORAGE AND RESUSPENSION Olfactory receptors are G protein-coupled receptor proteins that localize to Store lyophilized siRNA duplex at -20° C with desiccant. Stable for at least the cilia of olfactory sensory neurons where they display affinity for and bind one year from the date of shipment. Once resuspended, store at -20° C, to a variety of odor molecules. The genes encoding olfactory receptors com- avoid contact with RNAses and repeated freeze thaw cycles. prise the largest family in the human genome. The binding of olfactory recep- Resuspend lyophilized siRNA duplex in 330 µl of the RNAse-free water tor proteins to odor molecules triggers a signal transduction cascade that provided. Resuspension of the siRNA duplex in 330 µl of RNAse-free water leads to the production of cAMP via an olfactory-enriched adenylate cyclase. makes a 10 µM solution in a 10 µM Tris-HCl, pH 8.0, 20 mM NaCl, 1 mM This event ultimately leads to transmission of action potentials to the brain EDTA buffered solution. and the subsequent perception of smell. OR3A2 (olfactory receptor 3A2), also known as OLFRA04, OR17-228 (olfactory receptor 17-228) or olfactory recep- APPLICATIONS tor OR17-14, is a 321 amino acid multi-pass membrane protein that belongs to the G protein-coupled receptor 1 family and functions as an odorant recep- OR3A2 siRNA (h) is recommended for the inhibition of OR3A2 expression in tor. The gene encoding OR3A2 maps to human chromosome 17p13.3. human cells. REFERENCES SUPPORT REAGENTS 1. -
Supplementary Tables 1
Supplementary Information No. Description Page No. 1. Supplementary Table 1. List of TCGA samples used in analyses 2 2. Supplementary Figure S1. Identification of EMT target genes 6 3. Supplementary Table 2. List of EMT-TF targets and LPA-S1P signaling 7 components 4. Supplementary Figure S2. Tumor classification by M-classification approach 8 5. Supplementary Figure S3. Derivation of the 39 metastases-associated gene 9 set 6. Supplementary Table 3. Literature based functional assignment of the 39 10 gene set with metastases 7. Supplementary Data1. Weighted gene co-expression network analysis 11-13 (WGCNA) and module identification (W-classification) 8. Supplementary Data 2. Classification and module gene validation in additional 14-17 Ovarian Cancer datasets (VDs) 9. Supplementary Data 3. Identification of class-defining features using Gene 18-20 Set Enrichment Analysis (GSEA) 10. Supplementary Table 4: Modules and associated Genes 21-42 11. Supplementary Table 5: Class Functionalities based on Enriched Module 43 Genes 12. Supplementary Table 6. Ovarian cancer cell line datasets 44 13. Supplementary Fig. S4. Representative Wound Healing Assay Images 45 14. Supplementary Fig. S5. Graphical representation of wound healing 46 15. Supplementary Figure S6. Representative immunofluorescence image of 46 Class2 invasion 16. Supplementary Figure S7. Representative Immunofluorescence expression 47 images of markers 17. Supplementary Table 7. Correlation between TCGA subtypes and MW 47 classes 18. SupplementaryTable 8. Correlation between MW classes -
Multi-Tissue Probabilistic Fine-Mapping of Transcriptome-Wide Association Study Identifies Cis-Regulated Genes for Miserableness
bioRxiv preprint doi: https://doi.org/10.1101/682633; this version posted June 26, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Multi-tissue probabilistic fine-mapping of transcriptome-wide association study identifies cis-regulated genes for miserableness Calwing Liao1,2 BSc, Veikko Vuokila2, Alexandre D Laporte2 BSc, Dan Spiegelman2 MSc, Patrick A. Dion2,3 PhD, Guy A. Rouleau1,2,3 * MD, PhD 1Department oF Human Genetics, McGill University, Montréal, Quebec, Canada 2Montreal Neurological Institute, McGill University, Montréal, Quebec, Canada 3Department oF Neurology and Neurosurgery, McGill University, Montréal, Quebec, Canada Short summary: The First transcriptome-wide association study oF miserableness identiFies many genes including c7orf50 implicated in the trait. Word count: 1,522 excluding abstract and reFerences Tables: 3 Keywords: Miserableness, transcriptome-wide association study, TWAS *Correspondence: Dr. Guy A. Rouleau Montreal Neurological Institute and Hospital Department oF Neurology and Neurosurgery 3801 University Street, Montreal, QC Canada H3A 2B4. Tel: +1 514 398 2690 Fax: +1 514 398 8248 E-mail: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/682633; this version posted June 26, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract (141 words) Miserableness is a behavioural trait that is characterized by strong negative Feelings in an individual. -
Explorations in Olfactory Receptor Structure and Function by Jianghai
Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 ABSTRACT Explorations in Olfactory Receptor Structure and Function by Jianghai Ho Department of Neurobiology Duke University Date:_______________________ Approved: ___________________________ Hiroaki Matsunami, Supervisor ___________________________ Jorg Grandl, Chair ___________________________ Marc Caron ___________________________ Sid Simon ___________________________ [Committee Member Name] An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Neurobiology in the Graduate School of Duke University 2014 Copyright by Jianghai Ho 2014 Abstract Olfaction is one of the most primitive of our senses, and the olfactory receptors that mediate this very important chemical sense comprise the largest family of genes in the mammalian genome. It is therefore surprising that we understand so little of how olfactory receptors work. In particular we have a poor idea of what chemicals are detected by most of the olfactory receptors in the genome, and for those receptors which we have paired with ligands, we know relatively little about how the structure of these ligands can either activate or inhibit the activation of these receptors. Furthermore the large repertoire of olfactory receptors, which belong to the G protein coupled receptor (GPCR) superfamily, can serve as a model to contribute to our broader understanding of GPCR-ligand binding, especially since GPCRs are important pharmaceutical targets. -

Online Supporting Information S2: Proteins in Each Negative Pathway
Online Supporting Information S2: Proteins in each negative pathway Index Proteins ADO,ACTA1,DEGS2,EPHA3,EPHB4,EPHX2,EPOR,EREG,FTH1,GAD1,HTR6, IGF1R,KIR2DL4,NCR3,NME7,NOTCH1,OR10S1,OR2T33,OR56B4,OR7A10, Negative_1 OR8G1,PDGFC,PLCZ1,PROC,PRPS2,PTAFR,SGPP2,STMN1,VDAC3,ATP6V0 A1,MAPKAPK2 DCC,IDS,VTN,ACTN2,AKR1B10,CACNA1A,CHIA,DAAM2,FUT5,GCLM,GNAZ Negative_2 ,ITPA,NEU4,NTF3,OR10A3,PAPSS1,PARD3,PLOD1,RGS3,SCLY,SHC1,TN FRSF4,TP53 Negative_3 DAO,CACNA1D,HMGCS2,LAMB4,OR56A3,PRKCQ,SLC25A5 IL5,LHB,PGD,ADCY3,ALDH1A3,ATP13A2,BUB3,CD244,CYFIP2,EPHX2,F CER1G,FGD1,FGF4,FZD9,HSD17B7,IL6R,ITGAV,LEFTY1,LIPG,MAN1C1, Negative_4 MPDZ,PGM1,PGM3,PIGM,PLD1,PPP3CC,TBXAS1,TKTL2,TPH2,YWHAQ,PPP 1R12A HK2,MOS,TKT,TNN,B3GALT4,B3GAT3,CASP7,CDH1,CYFIP1,EFNA5,EXTL 1,FCGR3B,FGF20,GSTA5,GUK1,HSD3B7,ITGB4,MCM6,MYH3,NOD1,OR10H Negative_5 1,OR1C1,OR1E1,OR4C11,OR56A3,PPA1,PRKAA1,PRKAB2,RDH5,SLC27A1 ,SLC2A4,SMPD2,STK36,THBS1,SERPINC1 TNR,ATP5A1,CNGB1,CX3CL1,DEGS1,DNMT3B,EFNB2,FMO2,GUCY1B3,JAG Negative_6 2,LARS2,NUMB,PCCB,PGAM1,PLA2G1B,PLOD2,PRDX6,PRPS1,RFXANK FER,MVD,PAH,ACTC1,ADCY4,ADCY8,CBR3,CLDN16,CPT1A,DDOST,DDX56 ,DKK1,EFNB1,EPHA8,FCGR3A,GLS2,GSTM1,GZMB,HADHA,IL13RA2,KIR2 Negative_7 DS4,KLRK1,LAMB4,LGMN,MAGI1,NUDT2,OR13A1,OR1I1,OR4D11,OR4X2, OR6K2,OR8B4,OXCT1,PIK3R4,PPM1A,PRKAG3,SELP,SPHK2,SUCLG1,TAS 1R2,TAS1R3,THY1,TUBA1C,ZIC2,AASDHPPT,SERPIND1 MTR,ACAT2,ADCY2,ATP5D,BMPR1A,CACNA1E,CD38,CYP2A7,DDIT4,EXTL Negative_8 1,FCER1G,FGD3,FZD5,ITGAM,MAPK8,NR4A1,OR10V1,OR4F17,OR52D1,O R8J3,PLD1,PPA1,PSEN2,SKP1,TACR3,VNN1,CTNNBIP1 APAF1,APOA1,CARD11,CCDC6,CSF3R,CYP4F2,DAPK1,FLOT1,GSTM1,IL2 -

Characterization of the Genomic Features and Expressed Fusion Genes In
1 SUPPLEMENTARY INFORMATION (ONLINE SUPPORTING INFORMATION) Characterization of the genomic features and expressed fusion genes in micropapillary carcinomas of the breast Natrajan et al. Supplementary Methods Supplementary Figures S1-S6 Supplementary Tables S1-S7 2 SUPPLEMENTARY METHODS Tumor samples Two cohorts of micropapillary carcinomas (MPCs) were analyzed; the first cohort comprised 16 consecutive formalin fixed paraffin embedded (FFPE) MPCs, 11 pure and 5 mixed, which were retrieved from the authors' institutions (Table 1), and a second, validation cohort comprised 14 additional consecutive FFPE MPCs, retrieved from Molinette Hospital, Turin, Italy. Frozen samples were available from five out of the 16 cases from the first cohort of MPCs. As a comparator for the results of the Sequenom mutation profiling, a cohort of 16 consecutive IC-NSTs matched to the first cohort of 16 MPCs according to ER and HER2 status and histological grade were retrieved from a series of breast cancers previously analyzed by aCGH[1]. In addition, 14 IC-NSTs matched according to grade, and ER and HER2 status to tumors from the second cohort of 14 MPCs, and 48 grade 3 IC-NSTs were retrieved from Hospital La Paz, Madrid, Spain[1] (Supplementary Table S1). Power calculation For power calculations, we have assumed that if MPCs were driven by a recurrent fusion gene in a way akin to secretory carcinomas (which harbor the ETV6-NTRK3 fusion gene in >95% of cases[2-4]) or adenoid cystic carcinomas of the breast (which harbor the MYB-NFIB fusion gene in >90% of cases[5]), a ‘pathognomonic’ driver event would be present in at least ≥70% of cases (an estimate that is conservative). -
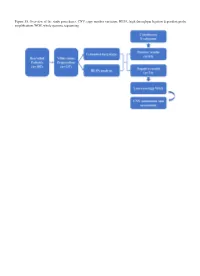
HLPA, High‑Throughput Ligation‑Dependent Probe Amplification; WGS, Whole‑Genome Sequencing
Figure S1. Overview of the study procedures. CNV, copy number variation; HLPA, high‑throughput ligation‑dependent probe amplification; WGS, whole‑genome sequencing. Table SI. Heredity tested by Sanger sequencing of NGS‑detected CNVs in abortion tissues. Heredity of CNVs Case CNV ‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑‑ Clinical number CNV location length (kb) Duplication/deletion Genes involved Paternal Maternal de novo significance V130 9p24.3 334 Duplication CBWD1, WASH1, FOXD4, DOCK8, FAM138C, C9orf66, ● VOUS DDX11L5 V134 3q13.13 504 Duplication ‑ ● Suspected benign 11q13.2 242 Deletion FAM86C2P ● VOUS 15q11.2 203 Deletion ‑ ● Suspected benign V138 2p24.3 811 Duplication ‑ ● Suspected benign V150 4q13.2 105 Deletion UGT2B28 ● VOUS 4q31.21 108 Duplication GYPB, GYPA ● VOUS V1001 11p15.1 320 Duplication TPH1, KCNC1, SERGEF ● VOUS 11q13.2 242 Deletion FAM86C2P ● VOUS 12p13.33 376 Duplication CACNA1C, LRTM2, CACNA2D4, LOC100271702, DCP1B ● VOUS V62 10q26.3 128 Deletion LOC619207, CYP2E1, SYCE1 ● VOUS 13q22.2 321 Deletion TBC1D4, COMMD6, UCHL3, LMO7 ● VOUS V114 4p15.2 191 Duplication C4orf52 ● VOUS V133 15q11.2 130 Deletion PWRN2 ● VOUS Xq27.3 202 Deletion ‑ ● Suspected benign V158 7q36.1 308 Duplication ATP6V0E2‑AS1, ZNF862, ATP6V0E2, DPP6 ● VOUS 7q36.2 183 Duplication LRP5L, CRYBB2P1, IGLL3P ● VOUS 22q11.23q12.1 263 Duplication ‑ ● VOUS V128 5q23.1 519 Duplication ‑ ● Suspected benign 22q11.21 270 Duplication PRODH, DGCR6, USP18, GGT3P ● VOUS Xp11.23 107 Deletion ZNF630, SPACA5B, SPACA5, SSX6 ● VOUS V54 1p11.12 104 Duplication -

Supplementary Methods
doi: 10.1038/nature06162 SUPPLEMENTARY INFORMATION Supplementary Methods Cloning of human odorant receptors 423 human odorant receptors were cloned with sequence information from The Olfactory Receptor Database (http://senselab.med.yale.edu/senselab/ORDB/default.asp). Of these, 335 were predicted to encode functional receptors, 45 were predicted to encode pseudogenes, 29 were putative variant pairs of the same genes, and 14 were duplicates. We adopted the nomenclature proposed by Doron Lancet 1. OR7D4 and the six intact odorant receptor genes in the OR7D4 gene cluster (OR1M1, OR7G2, OR7G1, OR7G3, OR7D2, and OR7E24) were used for functional analyses. SNPs in these odorant receptors were identified from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) or through genotyping. OR7D4 single nucleotide variants were generated by cloning the reference sequence from a subject or by inducing polymorphic SNPs by site-directed mutagenesis using overlap extension PCR. Single nucleotide and frameshift variants for the six intact odorant receptors in the same gene cluster as OR7D4 were generated by cloning the respective genes from the genomic DNA of each subject. The chimpanzee OR7D4 orthologue was amplified from chimpanzee genomic DNA (Coriell Cell Repositories). Odorant receptors that contain the first 20 amino acids of human rhodopsin tag 2 in pCI (Promega) were expressed in the Hana3A cell line along with a short form of mRTP1 called RTP1S, (M37 to the C-terminal end), which enhances functional expression of the odorant receptors 3. For experiments with untagged odorant receptors, OR7D4 RT and S84N variants without the Rho tag were cloned into pCI. -

( 12 ) United States Patent
US010209239B1 (12 ) United States Patent (10 ) Patent No. : US 10 , 209 , 239 B1 Hanson et al . (45 ) Date of Patent: Feb . 19 , 2019 (54 ) METHOD OF MAKING AN AROMAGRAPH 7 , 122 ,305 B2 10 /2006 Klein et al. 7 ,223 ,533 B2 5 / 2007 Ostanin et al . COMPRISING ECTOPIC OLFACTORY 7 , 223 ,550 B2 5 / 2007 Dhanasekaran et al. RECEPTORS 7 , 235 ,648 B1 6 / 2007 Fowlkes et al . 7 , 250 , 263 B2 7 / 2007 Klein et al. (71 ) Applicant: Aromyx Corporation , Palo Alto , CA 7 , 273 ,747 B2 9 / 2007 Manfredi et al . (US ) 7 , 319 ,009 B2 1 / 2008 Klein et al. 7 , 361, 498 B2 4 / 2008 Fowlkes et al. 7 ,416 , 881 B1 8 / 2008 Fowlkes et al . ( 72 ) Inventors : Chris Hanson , Sunnyvale , CA (US ) ; 7 ,425 , 445 B2 9 /2008 Matsunami et al. William Harries , Boulder Creek , CA 7 ,611 , 854 B2 11/ 2009 Fowlkes et al. (US ) 7 ,691 , 592 B2 4 / 2010 Matsunami et al . 7 , 838 , 288 B2 11/ 2010 Matsunami et al. ( 73 ) Assignee : Aromyx Corporation , Palo Alto , CA 7 , 879 , 565 B2 2 / 2011 Matsunami et al . 8 ,298 ,781 B2 10 / 2012 Matsunami et al. (US ) 9 ,611 , 308 B2 4 /2017 Matsunami et al . 2008 /0299586 Al 12 /2008 Han et al . ( * ) Notice: Subject to any disclaimer, the term of this 2012 /0021932 AL 1 /2012 Mershin et al. patent is extended or adjusted under 35 2012 /0077210 A1 * 3 /2012 Trowell .. .. .. .. .. GOIN 33 /542 U . S . C . 154 (b ) by 0 days . 435 / 7 . 9 2013/ 0216492 AL 8 / 2013 Kato et al.