Population Structure and Genetic Bottleneck Analysis of Ankleshwar Poultry Breed by Microsatellite Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DTSI, Rajpipla
GUJARAT COUNCIL OF VOCATIONAL TRAINING 3rd floor, Block No. 8, Dr. Jivraj Mehta Bhavan Ghandhinagar Name of Exam 901 - Course on Computer Concept (CCC) Page no : 1 Center Name : 801 - DTSI RAJPIPLA Date of Exam 19/11/2008 Candidate Name and Designation Practical / Seat No Result Department Theory Marks Training Period Direct Exam 1 TADVI NARANBHAI JESANGBHAI Sr.No 22 34 ARM HEAD CONSTABLE 80190122022 12 Fail OFFICE OF THE SUPDTOF POLICE 2 PATEL MAHENDRAKUMAR VITTHALBHAI Sr.No 28 40 PRINCIPAL 80190122041 12 Fail SHREE VALLABH VIDYA NIKETAN BHADAM 3 PATEL DIVYANGKUMAR JAYANTILAL Sr.No 30 59 SATHI SAHAYAK 80190122042 29 Pass SHREE VALLABH VIDYA NIKETAN BHADAM 4 VASAVA HARIYABHAI CHIMANBHAI Sr.No 29 57 PEON 80190122043 28 Pass SHREE VALLABH VIDYA NIKETAN BHADAM 5 PATEL MRUDULABEN DINUBHAI Sr.No 27 57 SHIKSHAN SAHAYAK 80190122045 30 Pass SHREE VALLABH VIDYA NIKETAN BHADAM 6 ZALA JITENDRASINH UMEDSINH Sr.No 27 53 ASSISTANT TEACHER 80190122046 26 Pass SHREE VALLABH VIDYA NIKETAN BHADAM 7 ATODARIYA NILAMKUMARI TAKHATSINH Sr.No Ab 0 PRIMARY TEACHER 80190122047 Ab Absent SHREE ADARSH KUMAR SHALA 8 ROHIT VIPULBHAI CHANDUBHAI Sr.No Ab 0 PRIMARY TEACHER 80190122048 Ab Absent PRATHMIK SHALA KAPAT 9 VASAVA ANANDBHAI BHANGABHAI Sr.No 26 53 CRC CO ORDINATOR 80190122049 27 Pass PRATHMIK SHALA CHKDA 10 PRAJAPATI ISHWARBHAI RAYAJIBHAI Sr.No 28 49 TEACHER 80190122050 21 Fail DANDIA BAZAR MISHRA SHALA 11 CHAUHAN DAKXABEN RANJITSINH Sr.No 25 50 CLERK 80190122051 25 Pass COLLECTOR OFFICE 12 VASAVA RAJANBHAI DHIRUBHAI Sr.No 25 52 CLERK 80190122052 27 Pass COLLECTOR OFFICE, GUJARAT COUNCIL OF VOCATIONAL TRAINING 3rd floor, Block No. -

Organic Farmers and Farms in Gujarat
Organic Farmers and Farms in Gujarat BHASKER SAVE’S KALPAVRUKSHA FARM Po : Deheri, Taluka : Umbergaon, District : Valsad – 396 170, Gujarat. Phone No. 0260-2562126 ‘Su-Swagatam’, proclaims a bright blue plaque with white lettering at the gate of Bhaskar Save’s verdant 14 acre farm, Kalpavruksha. About twenty steps from the gate is another sign that says: ‘Co-operation is the fundamental law of Nature’. Further inside are numerous other sign-boards that attract attention with brief, thought-provoking ‘sutras’ or aphorisms. These pithy sayings contain all the distilled wisdom on nature, farming, health, culture and spirituality that Bhaskar bhai has gathered over the years, apart from his extraordinary harvest of food. If you ask this warm humble farmer where he learnt his way of natural farming, he might tell you, ‘My university is my farm.’ And now, his farm has become a sacred university for many, as every Saturday afternoon brings a few dozen or more visitors. Included in the entourage are farmers, agricultural scientists, students, city folk, government officials, VIPs and the occasional foreigner who has read or heard of Bhaskar Save’s work. Kalpavruksha compels attention. For, its high yield easily out-performs any modern farm using chemicals. This is readily visible. The number and quality of the coconuts per tree are among the highest in the country. Some of the palms yield over 500 coconuts each year, while the average is about 400. The crop of chikoo (sapota) is similarly abundant and of excellent flavour providing on average 300-350 kg premium quality fruit per tree each year. -

A) APPLICATION for TERMS of REFERENCES (Tor
M/s Shree Narmada Khand Udyog Sahakari Mandli Ltd., Dharikheda, Tal.- Rajpipla (Nandod), Dist.- Narmada, Gujarat (A) APPLICATION FOR TERMS OF REFERENCES (ToR) Page 1 M/s Shree Narmada Khand Udyog Sahakari Mandli Ltd., Dharikheda, Tal.- Rajpipla (Nandod), Dist.- Narmada, Gujarat (B) FORM 1 SUBMITTED FOR ToR Page 2 M/s Shree Narmada Khand Udyog Sahakari Mandli Ltd., Dharikheda, Tal.- Rajpipla (Nandod), Dist.- Narmada, Gujarat APPENDIX I (See paragraph – 6) FORM 1 Sr. Item Details No. M/s Shree Narmada Khand Udyog 1 Name of the Project/s SahakriMandli Ltd., A/p – Dharikheda, Tal: Rajpipla (Nandod),Dist.: Narmada, Gujarat 5 (g) as per the provision of “EIA 2 S. No in the Schedule Notification No. S. O. 1533 (E)” dated 14.09.2006 amended on January 19, 2009. Proposed capacity / area / length / tonnage to be Products Capacity 3 handled / command area / lease area / number of Molasses based Distillery 45 KLPD wells to be drilled Unit It is a Proposed Distillery Unit 4 New / Expansion / Modernization 5 Existing capacity / Area etc. Existing Sugar Plant of capacity 2500 TCD As per the provision of “EIA 6 Category of Project i.e. ‘A’ or ‘B’ Notification No. S. O. 1533 (E)” dated 14.09.2006 amended on January 19, 2009 the proposed project comes under Category ‘A’. Does it attract the general condition? 7 Not Applicable If Yes please Specify. Does it attract the specific condition? 8 Not Applicable If yes please specify. Location Latitude – 21.827174 0N, Longitude – 73.388085 0E Refer prefeasibility report for Google Image 9 Plot/survey/Khasra No. -

Interview List for Selection of Appointment of Notaries in the State of Gujarat
Interview List for Selection of Appointment of Notaries in the State of Gujarat Area of Practice S.No. Name File No. Father Name Address Enrollment no. Applied for 2, ManubhailS Chawl, Nisarahmed N- Ahmedabad Gulamrasul Near Patrewali Masjid G/370/1999 1 Gulamrasul 11013/2011/2016- Metropolitan City A.Samad Ansari Gomtipur Ahmedabad Dt.21.03.99 Ansari NC Gujarat380021 N- Gulamnabi At & Po.Anand, B, Nishant Ms. Merunisha G/1267/1999 2 Anand Distt. 11013/2012/2016- Chandbhai Colony, Bhalej Road, I Gulamnabi Vohra Dt.21.03.99 NC Vohra Anand Gujarat-388001 333, Kalpna Society, N- Deepakbhai B/H.Suryanagar Bus G/249/1981 3 Vadodara Distt. 11013/2013/2016- Bhikubhai Shah Bhikubhai Shah Stand, Waghodia Road, Dt.06.05.81 NC Vadodara Gujarat- N- Jinabhai Dhebar Faliya Kundishery Ms. Bakula G/267/1995 4 Junagadh 11013/2014/2016- Jesabhai Arunoday Junagadh Jinabhai Dayatar Dt.15.03.95 NC Dayatar Dist.Junagadh Gujarat- Mehta N- Vill. Durgapur, Tal. G/944/1999 5 Bharatkumar Mandvi-Kutch 11013/2015/2016- Hirji Ajramal Manvdi-Kutch Gujarat Dt.21.03.99 Hirji NC N- At.Kolavna, Ta.Amod, Patel Mohamedali G/857/1998 6 Bharuch Distt. 11013/2016/2016- Patel Yakub Vali Distt.Bharuch, Gujarat- Yakub Dt.09.10.98 NC 392140 N- Gandhi 6-B/1, Ajay Wadi, Gandhi Hitesh G/641/2000 7 Bhavnagar 11013/2017/2016- Vasantray Subhashnagar Bhavnagar Vasantray Dt.05.05.2000 NC Prabhudas Gujarat- 319, Suthar Faliyu, At. & N- Nileshkumar Jhagadiya, Dist. Motibhai PO Avidha, Ta. Jhagadiya, G/539/1995 8 11013/2018/2016- Motibhai Desai Bharuch Laxmidas Desai Dist. -
Narmada District
Government of India Ministry of Water Resources GROUNDWATER BROCHURE NARMADA DISTRICT Compiled By H N Tiwari Scientist – C Central Ground Water Board West Central Region Ahmedabad March 2014 DISTRICT GROUND WATER BROCHURE NARMADA DISTRICT, GUJARAT INTRODUCTION Narmada district is situated in the southern part of Gujarat State. The district lies between north latitudes 21°23’ and 22°05’ and east longitudes 73°17’ and 73°59’. The geographical area of the district is 2755 sq.km. Narmada district is bounded in the North by Vadodara district, in the south by Surat district, in the west by Bharuch district and in east by Maharashtra State (Fig.1). This district was carved out on October 2, 1997. The newly formed district consist of Tilakwada taluka of erstwhile Vadodara district and 3 talukas viz., Nandod, Dediyapada and Sagbara of erstwhile Bharuch district. and consists of 609 villages. Total population of the district as per 2011 census is 5,90,379. Fig-1 STUDIES/ACTIVITIES BY CGWB This district was carved out on October 2, 1997. The newly formed district consist of Tilakwada taluka of erstwhile Vadodara district and 3 talukas viz., Nandod, Dediyapada and Sagbara of erstwhile Bharuch district. Therefore most of the previous work done in the Bharuch district and Varodara district is now covered in Narmada district. A. R. Pandey of Central Ground Water Board (1977-78) carried out detailed hydrogeological investigation in parts of Bharuch distrlict. Sporadic hydrogeological survey work in connection with drilling and construction of tube wells in parts of Bharuch district was carried out by J. S. -

Permission for Laying Proposed 125 Mm Dia PE Gas Pipeline Along
eerie ont Government of India Ministry of Road Transport & Highways (Regional Office, Gandhinagar, Dadra& Nagar Haveli and Daman& Diu) 4 floor, Block No. 14, New Sachivalaya, Regional Office, Gandhinagar, Gujarat-382010 Phone No. 079-26950705, Phone/Fax No.079-26950705 [email protected] RW/GNR/NH/US/GL/NOC/217/2020 | 161 Date: 24 March 2021 , Invitation of Public Comments Subject: Permission for laying proposed 125 mm dia PE gas pipeline along/across NH-56 (Rajpipla-Netrang) & (Rajpipla -Devaliya) to set up city Gas Distribution Network in Narmada GA in the state of Gujarat. Reference: Letter No. NHP-18-2021-189-M dated 06.02.2021 The Under Secretary , Roads and Building Department, Government of Gujarat vide letter cited under reference has recommended the subject cited proposal proposed 125 mm dia PE gas pipeline along/across NH-56 (Rajpipla-Netrang) & (Rajpipla -Devaliya) to set up city Gas Distribution Network in Narmada GA in the state of Gujarat for approval of Regional Officer , MoRT&H Gujarat. Zi As per Ministry vide OM No. RW/NH-33044/29/2015/S&R(R) dated 22.11.2016, the Highway Administration will make available the proposal seeking permission for utility laying for public comments for 30 days on ground of general public interest for seeking claims and objections. 3. In view of the above the comments of public are invited on captioned proposal and the same should reach to below mentioned address till 26.04.2021 beyond which no comments will be considered. The Regional officer, Gandhinagar Ministry of Road Transport & Highways (Regional Office, Gandhinagar, Dadra& Nagar Haveli and Daman& Diu) Ath floor, Block No. -

Bharuch District Disaster Management Plan 2019-2020 &
Bharuch District Disaster Management Plan 2019 -2020 Collector Office - Bharuch & Gujarat State Disaster Management Authority 1 DDMP- BHARUCH 2019-20 BHARUCH DISTRICT DISASTER MANAGEMENT PLAN Name of the District : - Bharuch Date (Plan Last Submitted) : - May 2018 Date (Plan Last Updated) :- May 2019 Signature of District Collector :- Emergency Operation Center, Collector Office, Bharuch & Gujarat State Disaster Management Authority May 2019 2 DDMP- BHARUCH 2019-20 Shri Ravi Kumar Arora (IAS) PREFACE The district disaster management plan is part of multi- level planning advocated by the Disaster Risk Management Programme an initiative of the government of Gujarat Preparedness of such a plan for Bharuch assumes significance given multi hazards scenario of the district i.e. natural and Disaster includes flood, cyclone, and storm surge and man-made includes industrial- chemical, fire, building collapse, communal right are main hazards of the district. Several efforts have been made over the years to address all these hazards by the district administration. However there remained several gaps which needed urgent plugging in. The Multi –Hazards District Disaster Management Plan of Bharuch District is basically action plan for various hazards, which envisages an Incident Response System with a clear line of command but also provide garter role clarity with delegation of specific power and responsibility to each staff in the structure This is a comprehensive document covering all possible hazards, with a systematic analysis of risk & vulnerability, elements at risk and level of compact through scientific and experience developed by the team of district administration. I appreciate the work carried out by Mr.shaileshkumar jiyani, District Project Officer of GSDMA and Bharuch district who has been taken lead for developing and compiling DDMP for the year 2018-2019. -

Assessment of Ground Water Quality in and Around Lignite Mine of Rajpardi, Bharcuh, Gujarat C
Mistry et al RJLBPCS 2019 www.rjlbpcs.com Life Science Informatics Publications Original Research Article DOI: 10.26479/2019.0501.13 ASSESSMENT OF GROUND WATER QUALITY IN AND AROUND LIGNITE MINE OF RAJPARDI, BHARCUH, GUJARAT C. P. Mistry1*, N. Sharma2, M. H. Trivedi1 1. Department of Earth and Environmental Science, KSKV Kachchh University, Mudra Road, Bhuj – 01, Gujarat, India. 2.Department of Geology, Faculty of Science, MS University of Baroda, Vadodara, Gujarat, India. ABSTRACT: Rajpardi lignite mine project, run by Gujarat Mineral Development Corporation, Ltd, Ahmedabad since 1983. Rajpardi is located in south Gujarat near Bharuch district. Production rate of lignite from the mine is approximately 10,00,000 tons per annum since its inception. Therefore, the impact of mining on ground water is likely to happen. In accordance, the present study was undertaken to assess the quality of ground water with respect to utility as portable and irrigation in farm land. Total 14 samples were collected in 10 km radius of mining area in triplicate for all three seasons i.e. winter, summer and post monsoon. Samples were brought to laboratory for the analysis of chemical and physical parameters. For computing the water budget, Gujarat Water Resource Development Corporation, Ltd., data is used. Quality of ground water is pristine except few sampling stations. High concentration of sodium and fluoride can cause impact like high blood pressure and fluorosis after long time consumption. In addition, presence of toxic substance like lead in sample of 1, 2, 3 and 14 are not recommended for drinking and irrigation purpose. KEYWORDS: Mining, lignite, water quality, heavy metals, water quality index. -
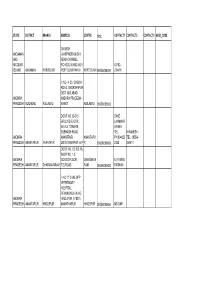
State District Branch Address Centre Ifsc Contact1 Contact2 Contact3 Micr Code
STATE DISTRICT BRANCH ADDRESS CENTRE IFSC CONTACT1 CONTACT2 CONTACT3 MICR_CODE CHURCH ANDAMAN LANEPHOENIX BAY AND NEAR CARAMEL NICOBAR SCHOOL WARD NO 6 03192- ISLAND ANDAMAN PORTBLAIR PORT BLAIR744101 PORT BLAIR BKID0008091 234400 H NO 4 -35 CINEMA ROAD BHOKTHAPUR DIST ADILABAD ANDHRA ANDHRA PRADESH - PRADESH ADILABAD ADILABAD 504001 ADILABAD BKID0005652 DOOR NO.12-313, SAKE GROUND FLOOR, LAXMINAR BALAJI TOWERS, AYANA, SUBHASH ROAD, TEL. K.RAMESH, ANDHRA ANANTPUR, ANANTAPU PH:994825 TEL. 08554- PRADESH ANANTAPUR ANANTPUR DIST:ANANTPUR, A.P R BKID0008689 2352 249411 DOOR NO. 12-100-1A, SHOP NO. 1-3, ANDHRA GOUND FLOOR, DHARMAVA K.THIMMA PRADESH ANANTAPUR DHARMAVARAM R.S.ROAD RAM BKID0005629 RATNAM H NO 17 3 645 OPP VETERINARY HOSPITAL PENUKONDA ROAD ANDHRA HINDUPUR 515201, PRADESH ANANTAPUR HINDUPUR ANANTHAPUR HINDUPUR BKID0005654 MD SAFI BANK OF INDIA, C. RAMAPURAM BRANCH, 4-1, GROUND FLOOR, C. RAMAPURAM VILLAGE, TIRUPATHI RURAL, CHITTOOR DIST., ANDHRA BR MGR ANDHRA PRADESH, PIN: 0877- PRADESH CHITTOOR C. RAMAPURAM 517561 CHITTOOR BKID0005718 2247096 IST FL.,MUNICIPAL SHOPPING COMPLEX, PRAKASAM HIGH SHRI N. G. ROAD, P.B.NO.13, REDDY, SHRI S. DIST. CHITTOOR, SR. SUBRAMANIA, ANDHRA MANAGER, SR. MANAGER, ANDHRA PRADESH.PIN 517 TEL:08572- TEL:08572- PRADESH CHITTOOR CHITOOR 001. CHITTOOR BKID0008670 233327 233327 D.NO-2-41,MAIN ROAD, KAYAMPETA- VILL,BRAHMANAPAT TU-PO, VADAMALPETA- ANDHRA MANDAL, CHITTOOR- VADAMALP 08577- 919959762 PRADESH CHITTOOR KAYAMPETA DIST, PIN-517551 ET BKID0005645 237929 RAMESH KVS 526 XVI 601 AND 608,SRI SAI,HARSHA COMPLEX,KAMMAPA LLI,EAST NIMMAPALI X ROAD, MADANAPALLI- ANDHRA 517325,CHITTOR MADANAPA 789332257 PRADESH CHITTOOR MADANAPALLI ANDHRA PRADESH LLE BKID0005646 0 7893322570 B-50/1, AIR BYPASS ROAD, OPP. -

Sidis in the Agate Bead Industry of Western India Kenoyer
Sdis and the Agate Bead Industry of Western India 61 Sidis and the Agate Bead Industry of Western India J Mark Kenoyer and Kuldeep K B1an Introduction The role of Gori Pir and the Sidi community in the history of Gujarat and western India is a long neglected topic. In this paper, historical records about the Sidis and their own oral traditions will be critically examined to gain new perspectives on their complex history, begin ning with their origins in Africa and with a special focus on their role in the agate bead industry. On the basis of current evidence it is clear that the Sidi communities of Gujarat and adjacent regions have made significant contributions to the larger sphere of South Asian culture. These contributions include agate mining and bead manufacture, as well as local trade and long distance trade networks that connected the Indian subcontinent to the furthest reaches of the Islamic world in Africa, Southeast Asia and West Asia. Who are the Sidi and Where did They Come From? These two questions can only be answered conclusively through care ful archaeological and DNA studies, but it is possible to make some sense out of the situation based on written documents and oral tradi tions. Some early anthropological studies of different racial groups in South Asia have shown that there are some Negrito groups among indigenous populations of hunter-gatherers living in parts ofthe West ern Ghats (Kadar and Irular of the Kerala Hills), and in the Raj Mahal Hills of Bihar and Bengal. However, except for the well known Negrito populations on the Andaman and Nicobar Islands, there is 62 Sdis and Silo/ars no conclusive evidence for indigenous Negrito populations on the mainland (Majumdar 1961: 52-53).