Data Management in Systems Biology I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Connectivity and Complex Systems: Learning from a Multi-Disciplinary Perspective
This is a repository copy of Connectivity and complex systems: learning from a multi-disciplinary perspective. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/141950/ Version: Published Version Article: Turnbull, L., Hütt, M-T., Ioannides, A.A. et al. (8 more authors) (2018) Connectivity and complex systems: learning from a multi-disciplinary perspective. Applied Network Science, 3 (11). ISSN 2364-8228 https://doi.org/10.1007/s41109-018-0067-2 Reuse This article is distributed under the terms of the Creative Commons Attribution (CC BY) licence. This licence allows you to distribute, remix, tweak, and build upon the work, even commercially, as long as you credit the authors for the original work. More information and the full terms of the licence here: https://creativecommons.org/licenses/ Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ Turnbull et al. Applied Network Science (2018) 3:11 https://doi.org/10.1007/s41109-018-0067-2 Applied Network Science REVIEW Open Access Connectivity and complex systems: learning from a multi-disciplinary perspective Laura Turnbull1* , Marc-Thorsten Hütt2, Andreas A. Ioannides3, Stuart Kininmonth4,5, Ronald Poeppl6, Klement Tockner7,8,9, Louise J. Bracken1, Saskia Keesstra10, Lichan Liu3, Rens Masselink10 and Anthony J. Parsons11 * Correspondence: Laura.Turnbull@ durham.ac.uk Abstract 1Durham University, Durham, UK Full list of author information is In recent years, parallel developments in disparate disciplines have focused on what available at the end of the article has come to be termed connectivity; a concept used in understanding and describing complex systems. -

Use of Bioinformatics Resources and Tools by Users of Bioinformatics Centers in India Meera Yadav University of Delhi, [email protected]
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Library Philosophy and Practice (e-journal) Libraries at University of Nebraska-Lincoln 2015 Use of Bioinformatics Resources and Tools by Users of Bioinformatics Centers in India meera yadav University of Delhi, [email protected] Manlunching Tawmbing Saha Institute of Nuclear Physisc, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/libphilprac Part of the Library and Information Science Commons yadav, meera and Tawmbing, Manlunching, "Use of Bioinformatics Resources and Tools by Users of Bioinformatics Centers in India" (2015). Library Philosophy and Practice (e-journal). 1254. http://digitalcommons.unl.edu/libphilprac/1254 Use of Bioinformatics Resources and Tools by Users of Bioinformatics Centers in India Dr Meera, Manlunching Department of Library and Information Science, University of Delhi, India [email protected], [email protected] Abstract Information plays a vital role in Bioinformatics to achieve the existing Bioinformatics information technologies. Librarians have to identify the information needs, uses and problems faced to meet the needs and requirements of the Bioinformatics users. The paper analyses the response of 315 Bioinformatics users of 15 Bioinformatics centers in India. The papers analyze the data with respect to use of different Bioinformatics databases and tools used by scholars and scientists, areas of major research in Bioinformatics, Major research project, thrust areas of research and use of different resources by the user. The study identifies the various Bioinformatics services and resources used by the Bioinformatics researchers. Keywords: Informaion services, Users, Inforamtion needs, Bioinformatics resources 1. Introduction ‘Needs’ refer to lack of self-sufficiency and also represent gaps in the present knowledge of the users. -

Establishing the Basis for a CIS (Computer Information Systems) Undergraduate Program: on Seeking the Body of Knowledge
Information Systems Education Journal (ISEDJ) 13 (5) ISSN: 1545-679X September 2015 Establishing the Basis for a CIS (Computer Information Systems) Undergraduate Program: On Seeking the Body of Knowledge Herbert E. Longenecker, Jr. [email protected] University of South Alabama Mobile, AL 36688, USA Jeffry Babb [email protected] West Texas A&M University Canyon, TX 79016, USA Leslie J. Waguespack [email protected] Bentley University Waltham, Massachusetts 02452, USA Thomas N. Janicki [email protected] University of North Carolina Wilmington Wilmington, NC 28403, USA David Feinstein [email protected] University of South Alabama Mobile, AL 36688, USA Abstract The evolution of computing education spans a spectrum from computer science (CS) grounded in the theory of computing, to information systems (IS), grounded in the organizational application of data processing. This paper reports on a project focusing on a particular slice of that spectrum commonly labeled as computer information systems (CIS) and reflected in undergraduate academic programs designed to prepare graduates for professions as software developers building systems in government, commercial and not-for-profit enterprises. These programs with varying titles number in the hundreds. This project is an effort to determine if a common knowledge footprint characterizes CIS. If so, an eventual goal would be to describe the proportions of those essential knowledge components and propose guidelines specifically for effective undergraduate CIS curricula. Professional computing societies (ACM, IEEE, AITP (formerly DPMA), etc.) over the past fifty years have sponsored curriculum guidelines for various slices of education that in aggregate offer a compendium of knowledge areas in ©2015 EDSIG (Education Special Interest Group of the AITP) Page 37 www.aitp-edsig.org /www.isedj.org Information Systems Education Journal (ISEDJ) 13 (5) ISSN: 1545-679X September 2015 computing. -
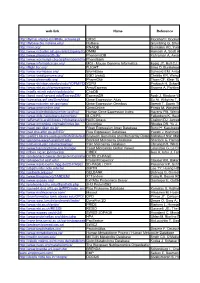
Web-Link Name Reference DRSC Flockhart I, Booker M, Kiger A, Et Al.: Flyrnai: the Drosophila Rnai Screening Center Database
web-link Name Reference http://flyrnai.org/cgi-bin/RNAi_screens.pl DRSC Flockhart I, Booker M, Kiger A, et al.: FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 34(Database issue):D489-94 (2006) http://flybase.bio.indiana.edu/ FlyBase Grumbling G, Strelets V.: FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34(Database issue):D484-8 (2006) http://rnai.org/ RNAiDB Gunsalus KC, Yueh WC, MacMenamin P, et al.: RNAiDB and PhenoBlast: web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 32(Database issue):D406-10 (2004) http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIMOMIM Hamosh A, Scott AF, Amberger JS, et al.: Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33(Database issue):D514-7 (2005) http://www.phenomicdb.de/ PhenomicDB Kahraman A, Avramov A, Nashev LG, et al.: PhenomicDB: a multi-species genotype/phenotype database for comparative phenomics. Bioinformatics 21(3):418-20 (2005) http://www.worm.mpi-cbg.de/phenobank2/cgi-bin/MenuPage.pyPhenoBank http://www.informatics.jax.org/ MGI - Mouse Genome Informatics Eppig JT, Bult CJ, Kadin JA, et al.: The Mouse Genome Database (MGD): from genes to mice--a community resource for mouse biology. Nucleic Acids Res. 33(Database issue):D471-5 (2005) http://flight.licr.org/ FLIGHT Sims D, Bursteinas B, Gao Q, et al.: FLIGHT: database and tools for the integration and cross-correlation of large-scale RNAi phenotypic datasets. Nucleic Acids Res. 34(Database issue):D479-83 (2006) http://www.wormbase.org/ WormBase Schwarz EM, Antoshechkin I, Bastiani C, et al.: WormBase: better software, richer content. -

The Design and Realisation of the Myexperiment Virtual Research Environment for Social Sharing of Workflows
The Design and Realisation of the myExperiment Virtual Research Environment for Social Sharing of Workflows David De Roure a Carole Goble b Robert Stevens b aUniversity of Southampton, UK bThe University of Manchester, UK Abstract In this paper we suggest that the full scientific potential of workflows will be achieved through mechanisms for sharing and collaboration, empowering scientists to spread their experimental protocols and to benefit from those of others. To facilitate this process we have designed and built the myExperiment Virtual Research Environ- ment for collaboration and sharing of workflows and experiments. In contrast to systems which simply make workflows available, myExperiment provides mecha- nisms to support the sharing of workflows within and across multiple communities. It achieves this by adopting a social web approach which is tailored to the par- ticular needs of the scientist. We present the motivation, design and realisation of myExperiment. Key words: Scientific Workflow, Workflow Management, Virtual Research Environment, Collaborative Computing, Taverna Workflow Workbench 1 Introduction Scientific workflows are attracting considerable attention in the community. Increasingly they support scientists in advancing research through in silico ex- perimentation, while the workflow systems themselves are the subject of ongo- ing research and development (1). The National Science Foundation Workshop on the Challenges of Scientific Workflows identified the potential for scientific advance as workflow systems address more sophisticated requirements and as workflows are created through collaborative design processes involving many scientists across disciplines (2). Rather than looking at the application or ma- chinery of workflow systems, it is this dimension of collaboration and sharing that is the focus of this paper. -

Information Systems Foundations Theory, Representation and Reality
Information Systems Foundations Theory, Representation and Reality Information Systems Foundations Theory, Representation and Reality Dennis N. Hart and Shirley D. Gregor (Editors) Workshop Chair Shirley D. Gregor ANU Program Chairs Dennis N. Hart ANU Shirley D. Gregor ANU Program Committee Bob Colomb University of Queensland Walter Fernandez ANU Steven Fraser ANU Sigi Goode ANU Peter Green University of Queensland Robert Johnston University of Melbourne Sumit Lodhia ANU Mike Metcalfe University of South Australia Graham Pervan Curtin University of Technology Michael Rosemann Queensland University of Technology Graeme Shanks University of Melbourne Tim Turner Australian Defence Force Academy Leoni Warne Defence Science and Technology Organisation David Wilson University of Technology, Sydney Published by ANU E Press The Australian National University Canberra ACT 0200, Australia Email: [email protected] This title is also available online at: http://epress.anu.edu.au/info_systems02_citation.html National Library of Australia Cataloguing-in-Publication entry Information systems foundations : theory, representation and reality Bibliography. ISBN 9781921313134 (pbk.) ISBN 9781921313141 (online) 1. Management information systems–Congresses. 2. Information resources management–Congresses. 658.4038 All rights reserved. No part of this publication may be reproduced, stored in a retrieval system or transmitted in any form or by any means, electronic, mechanical, photocopying or otherwise, without the prior permission of the publisher. Cover design by Brendon McKinley with logo by Michael Gregor Authors’ photographs on back cover: ANU Photography Printed by University Printing Services, ANU This edition © 2007 ANU E Press Table of Contents Preface vii The Papers ix Theory Designing for Mutability in Information Systems Artifacts, Shirley Gregor and Juhani Iivari 3 The Eect of the Application Domain in IS Problem Solving: A Theoretical Analysis, Iris Vessey 25 Towards a Unied Theory of Fit: Task, Technology and Individual, Michael J. -

Information System Owner
DOE CYBERSECURITY: CORE COMPETENCY TRAINING REQUIREMENTS Key Cybersecurity Role: Information System Owner Role Definition: The Information System Owner (also referred to as System Owner) is the individual responsible for the overall procurement, development, integration, modification, operation, maintenance, and retirement of an information system. The System Owner is a key contributor in developing system design specifications to ensure the security and user operational needs are documented, tested, and implemented. Competency Area: Data Security Functional Requirement: Design Competency Definition: Refers to the application of the principles, policies, and procedures necessary to ensure the confidentiality, integrity, availability, and privacy of data in all forms of media (i.e., electronic and hardcopy) throughout the data life cycle. Behavioral Outcome: The System Owner will understand the policies and procedures required to protect all categories of information as well as have a working knowledge of data access controls required to ensure the confidentiality, integrity, and availability of information. He/she will apply this knowledge during all phases of the System Development Life Cycle (SDLC). Training concepts to be addressed at a minimum: Identify and document the appropriate level of protection for data, including use of encryption. Specify data and information classification, sensitivity, and need-to-know requirements by information type on a system in terms of its confidentiality, integrity, and availability. Utilize DOE M 205.1-5 to determine the information impacts for unclassified information and DOE M 205.1-4 to determine the Consequence of Loss for classified information. Create authentication and authorization system for users to gain access to data based on assigned privileges and permissions. -

Data Curation+Process Curation^Data Integration+Science
BRIEFINGS IN BIOINFORMATICS. VOL 9. NO 6. 506^517 doi:10.1093/bib/bbn034 Advance Access publication December 6, 2008 Data curation 1 process curation^data integration 1 science Carole Goble, Robert Stevens, Duncan Hull, Katy Wolstencroft and Rodrigo Lopez Submitted: 16th May 2008; Received (in revised form): 25th July 2008 Abstract In bioinformatics, we are familiar with the idea of curated data as a prerequisite for data integration. We neglect, often to our cost, the curation and cataloguing of the processes that we use to integrate and analyse our data. Downloaded from https://academic.oup.com/bib/article/9/6/506/223646 by guest on 27 September 2021 Programmatic access to services, for data and processes, means that compositions of services can be made that represent the in silico experiments or processes that bioinformaticians perform. Data integration through workflows depends on being able to know what services exist and where to find those services. The large number of services and the operations they perform, their arbitrary naming and lack of documentation, however, mean that they can be difficult to use. The workflows themselves are composite processes that could be pooled and reused but only if they too can be found and understood. Thus appropriate curation, including semantic mark-up, would enable processes to be found, maintained and consequently used more easily.This broader view on semantic annotation is vital for full data integration that is necessary for the modern scientific analyses in biology.This article will brief the community on the current state of the art and the current challenges for process curation, both within and without the Life Sciences. -

Chemical and Systems Biology, See the “Chemical Systems Courtesy Professors: Stuart Kim, Beverly S
undergraduates. The limited size of the labs in the department allows for close tutorial contact between students, postdoctoral fellows, and faculty. CHEMICAL AND SYSTEMS The department participates in the four quarter Health and Human Disease and Practice of Medicine sequence which provides medical students with a comprehensive, systems-based education in BIOLOGY physiology, pathology, microbiology, and pharmacology. Emeriti: (Professors) Robert H. Dreisbach, Avram Goldstein, Dora B. Goldstein, Tag E. Mansour, Oleg Jardetzky, James P. Whitlock CHEMICAL AND SYSTEMS Chair: James E. Ferrell, Jr. Professors: James E. Ferrell, Jr., Tobias Meyer, Daria Mochly- Rosen, Richard A. Roth BIOLOGY (CSB) COURSES Associate Professor: Karlene A. Cimprich Assistant Professors: James K. Chen, Thomas J. Wandless, Joanna For information on graduate programs in the Department of K. Wysocka Chemical and Systems Biology, see the “Chemical Systems Courtesy Professors: Stuart Kim, Beverly S. Mitchell, Paul A. Biology” section of this bulletin. Course and laboratory instruction Wender in the Department of Chemical and Systems Biology conforms to the Courtesy Associate Professor: Calvin J. Kuo “Policy on the Use of Vertebrate Animals in Teaching Activities,” Courtesy Assistant Professors: Matthew Bogyo, Marcus Covert the text of which is available at Consulting Professor: Juan Jaen http://www.stanford.edu/dept/DoR/rph/8-2.html. Web Site: http://casb.stanford.edu Courses offered by the Department of Chemical and Systems UNDERGRADUATE COURSES IN CHEMICAL Biology have the subject code CSB, and are listed in the “Chemical AND SYSTEMS BIOLOGY and Systems Biology (CSB) Courses” section of this bulletin. CSB 199. Undergraduate Research In Autumn of 2006, the Department of Molecular Pharmacology Students undertake investigations sponsored by individual faculty changed its name to become the Department of Chemical and members. -

Information System Development Using Augmented Reality Tools*
Information system development using augmented reality tools* Grigorev Rostislav Aleksandrovich Valijanov Bahrom Adhamdzon-ugli Kazan Federal University Kazan Federal University Kazan, Russia Kazan, Russia [email protected] [email protected] Medvedeva Olga Anatolievna Mustafina Svetlana Anatol’evna Kazan Federal University Bashkir State University Kazan, Russia Ufa, Russia [email protected] [email protected] Abstract This work is devoted to the issues of visualization and information processing, in particular, to improving the visualization of 3D objects using augmented reality technology. The concept of augmented reality offers a more advanced user interface for visualization due to a combination of natural ways to control the change of angle of an object and visualization in a real context. In the process of performing the work, computer graphics, algorithms, and modeling methods were used. The experimental part of the work was carried out using a set of development tools for tracking Vuforia and Unity development tools. Introduction Augmented reality (AR) is an interactive experience of a real-world environment where the objects that reside in the real world are enhanced by computer-generated perceptual information, sometimes across multiple sensory modalities, including visual, auditory, haptic, somatosensory and olfactory. AR can be defined as a system that fulfills three basic features: a combination of real and virtual worlds, real-time interaction, and accurate 3D registration of virtual and real objects. The overlaid sensory information can be constructive (i.e. additive to the natural environment), or destructive (i.e. masking of the natural environment). This experience is seamlessly interwoven with the physical world such that it is perceived as an immersive aspect of the real environment. -

Online Identity in the Case of the Share Phenomenon. a Glimpse Into the on Lives of Romanian Millennials
Online identity in the case of the share phenomenon. A glimpse into the on lives of Romanian millennials Demetra GARBAȘEVSCHI PhD Student National University of Political Science and Public Administration E-Mail: [email protected] Abstract. In less than a decade, the World Wide Web has evolved from a predominantly search medium to a predominantly share medium, from holding a functional role to being endowed with a social one.In the context of a reontologisation of the infosphere and of an unprecedented display of mass self-communication, the identity system has gained a legitimate dimension – online identity –, as individuals have become the sum of impressions openly offered online and decoded into a coherent story by the receiver. In the network society, there are consequences to both having and not having an online identity. Originating in an interactionist perspective, the present paper looks into Romanian Millennials in trying to find out whether online identity is undergoing a process of intentionalization, in other words whether it becomes a conscious, planned effort of the individual to build himself/ herself a legitimate and profitable dimension in the digital space. Keywords: online identity; infosphere; mass self-communication; Millennials; Generation Y. 1. Introduction and theoretical background This paper examines online identity as part of an individual’s identity system, in the specific context of current Internet development, generalized connectivity Journal of Media Research, Vol. 8 Issue 2(22) / 2015,14 pp. 14-26 and participation through the share web. The discussion centers on Romanian young adults, seeking to uncover their perceptions of online identity as a potentially strategic self-representation process. -

A Plaidoyer for 'Systems Immunology'
Christophe Benoist A Plaidoyer for ‘Systems Immunology’ Ronald N. Germain Diane Mathis Authors’ address Summary: A complete understanding of the immune system will ulti- Christophe Benoist1, Ronald N. Germain2, Diane mately require an integrated perspective on how genetic and epigenetic Mathis1 entities work together to produce the range of physiologic and patho- 1Section on Immunology and Immunogenetics, logic behaviors characteristic of immune function. The immune network Joslin Diabetes Center, Department of encompasses all of the connections and regulatory associations between Medicine, Brigham and Women’s Hospital, individual cells and the sum of interactions between gene products Harvard Medical School, Boston, MA, USA within a cell. With 30 000+ protein-coding genes in a mammalian 2Lymphocyte Biology Section, Laboratory of genome, further compounded by microRNAs and yet unrecognized Immunology, National Institute of Allergy and layers of genetic controls, connecting the dots of this network is a Infectious Diseases, National Institutes of monumental task. Over the past few years, high-throughput techniques Health, Bethesda, MD, USA have allowed a genome-scale view on cell states and cell- or system-level responses to perturbations. Here, we observe that after an early burst of Correspondence to: enthusiasm, there has developed a distinct resistance to placing a high Christophe Benoist value on global genomic or proteomic analyses. Such reluctance has Department of Medicine affected both the practice and the publication of immunological science, Section on Immunology and Immunogenetics resulting in a substantial impediment to the advances in our understand- Joslin Diabetes Center ing that such large-scale studies could potentially provide. We propose Brigham and Women’s Hospital that distinct standards are needed for validation, evaluation, and visual- Harvard Medical School ization of global analyses, such that in-depth descriptions of cellular One Joslin Place responses may complement the gene/factor-centric approaches currently Boston, MA 02215 in favor.