DNA Minibarcodes in Taxonomic Assignment: a Morphologically
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Wildlife Matters: Winter 2016 1 Wildlife Matters
Wildlife Matters: Winter 2016 1 wildlife matters Winter 2016 Historic partnership: AWC to reintroduce lost mammals to NSW national parks 2 Wildlife Matters: Winter 2016 Saving Australia’s threatened wildlife Welcome to the Winter 2016 edition of Wildlife Matters. The AWC mission This edition marks the beginning of a historic partnership between Australian The mission of Australian Wildlife Wildlife Conservancy (AWC) and the NSW Government. AWC has been contracted Conservancy (AWC) is the effective to deliver national park management services in the iconic Pilliga forest and at conservation of all Australian animal Mallee Cliffs National Park in the state’s south-west. It is the first public-private species and the habitats in which they live. collaboration of its kind. The centrepiece of this exciting partnership will be the reintroduction of at least 10 mammal species that are currently listed as extinct in To achieve this mission our actions are NSW. focused on: This is one of the world’s most significant biodiversity reconstruction projects. The • Establishing a network of sanctuaries return of mammals such as the Bilby and the Numbat – which disappeared from which protect threatened wildlife and NSW national parks more than 100 years ago – will represent a defining moment in ecosystems: AWC now manages our quest to halt and reverse the loss of Australia’s unique wildlife. 25 sanctuaries covering over 3.25 million hectares (8 million acres). The initiative reflects strong leadership by the NSW Government. It is committing substantial funds for threatened species, including this partnership with AWC. • Implementing practical, on-ground More importantly, the NSW Government recognises the need to develop new conservation programs to protect approaches to conservation if we are to reverse the catastrophic decline of the wildlife at our sanctuaries: these Australia’s natural capital. -

Erection of a New Family in the Lepidopterous Suborder Dacnonypha
Entomologiske M eddelelser 35 (1967) 341 Erection of a New Family in the Lepidopterous Suborder Dacnonypha. By N. P. Kristensen Zoological Institute, University of Copenhagen. The homoneurous moth genus Jlgathiphaga was described by Dumbleton in 1952. The genus comprises two species, occuring in Queensland (Australia) and on Fiji, respectively; the larvae of both feed in the seeds of Kauri pines (Agathis). The adult ana tomy indicated affinities to both Micropterygidae and Eriocranii dae; Dumbleton decided however, that the weight of evidence was for considering Agathiphaga as a specialized genus of Micropte rygidae. On the other hand Hinton (1958) after examining the Agathiphaga-larvae found these to possess several apomorph characters characteristic of Dacnonypha and higher Lepidoptera and to be devoid of any of the features characteristic of the Micropterygid larvae. He therefore concluded that the genus belongs to the Eriocraniidae or a closely related family. The correctness of the transferring of Agathiphaga to the suborder Dacnonypha cannot be doubted; however, in the adult anatomy the genus differs from the Eriocraniidae as well as from the other dacnonyphous families (Mnesarchaeidae, Neopseustidae) in many important features, and consequently has to be regarded as con stituting a separate family, which is defined below. Agathiphagidae fam. nov. Type-genus: Agathiphaga Dumbleton, 1952. D i a g no si s. Adult: Articulated mandibles present, galeae nol haustellate, lobular lacinia present, tibia 2 and 3 with paired subapical and apical spurs, forewing with closed cell between M and Cu, d -genitalia with long and simple, dorsally curved valvae, phallus with short posteriorly directed ventral apodeme. 22* 342 N. -

Verbreitung Und Wissensstand Gerald Fuchs
Dieses PDF wird von der Arbeitsgemeinschaft bayerischer Entomologen e.V.für den privaten bzw. wissenschaftlichen Gebrauch zur Verfügung gestellt. Die kommerzielle Nutzung oder die Bereitstellung in einer öffentlichen Bibliothek oder auf einer website ist nicht gestattet. Beiträge zur bayerischen Entomofaunistik 14:5–24, Bamberg (2014), ISSN 1430-015X Die bayerischen Urmotten – Verbreitung und Wissensstand (Insecta: Lepidoptera: Micropterigidae) von Gerald Fuchs Abstract: In the German federal county of Bavaria 9 species of the lepidoptera family Micropterigidae are presently known. This paper provides an overview of their distribution and current scientific knowledge of the species in Bavaria. Zusammenfassung: In Bayern wurden bisher 9 Arten aus der Familie Micropterigidae nachgewiesen. Diese Arbeit gibt eine Übersicht über die bisher bekannt gewordene Verbreitung und den aktuellen Wissenstand der bayerischen Arten. Einleitung Während die Erfassung und Zusammenführung des großen Bestandes an bayerischen Daten zu den Tagfal- tern 2013 in eine Publikation mündete (Bräu et al., 2013), ist über die bayerische Gesamtverbreitung aller anderen Familien bisher nichts veröffentlicht worden. Die Familie Micropterigidae (Urmotten) soll hier eine erste Lücke schließen, auch wenn der Datenbestand auf einer relativ geringen Grundlage basiert, und dazu anregen, sich intensiver mit dieser Gruppe zu beschäftigen. Die Bestimmung der Individuen ist heutzutage recht einfach, da gute Literatur vorhanden ist (Zel- ler-Lukashort et al., 2007; Whitebread, 1990). Selbst abgeflogene männliche Tiere können ohne gro- ßen Präparationsaufwand aufgrund des ausgestülpten Genitals leicht determiniert werden. Bei abgeflo- genen Weibchen ist dies dafür umso schwieriger, da der Genitalapparat nicht oder nur sehr schwach sklerotisiert ist und sich somit nicht für die übliche Anfertigung von Dauerpräparaten eignet. In Zel- ler-Lukashort et al. -

Assessment of Forest Pests and Diseases in Protected Areas of Georgia Final Report
Assessment of Forest Pests and Diseases in Protected Areas of Georgia Final report Dr. Iryna Matsiakh Tbilisi 2014 This publication has been produced with the assistance of the European Union. The content, findings, interpretations, and conclusions of this publication are the sole responsibility of the FLEG II (ENPI East) Programme Team (www.enpi-fleg.org) and can in no way be taken to reflect the views of the European Union. The views expressed do not necessarily reflect those of the Implementing Organizations. CONTENTS LIST OF TABLES AND FIGURES ............................................................................................................................. 3 ABBREVIATIONS AND ACRONYMS ...................................................................................................................... 6 EXECUTIVE SUMMARY .............................................................................................................................................. 7 Background information ...................................................................................................................................... 7 Literature review ...................................................................................................................................................... 7 Methodology ................................................................................................................................................................. 8 Results and Discussion .......................................................................................................................................... -

Lepidoptera of North America 5
Lepidoptera of North America 5. Contributions to the Knowledge of Southern West Virginia Lepidoptera Contributions of the C.P. Gillette Museum of Arthropod Diversity Colorado State University Lepidoptera of North America 5. Contributions to the Knowledge of Southern West Virginia Lepidoptera by Valerio Albu, 1411 E. Sweetbriar Drive Fresno, CA 93720 and Eric Metzler, 1241 Kildale Square North Columbus, OH 43229 April 30, 2004 Contributions of the C.P. Gillette Museum of Arthropod Diversity Colorado State University Cover illustration: Blueberry Sphinx (Paonias astylus (Drury)], an eastern endemic. Photo by Valeriu Albu. ISBN 1084-8819 This publication and others in the series may be ordered from the C.P. Gillette Museum of Arthropod Diversity, Department of Bioagricultural Sciences and Pest Management Colorado State University, Fort Collins, CO 80523 Abstract A list of 1531 species ofLepidoptera is presented, collected over 15 years (1988 to 2002), in eleven southern West Virginia counties. A variety of collecting methods was used, including netting, light attracting, light trapping and pheromone trapping. The specimens were identified by the currently available pictorial sources and determination keys. Many were also sent to specialists for confirmation or identification. The majority of the data was from Kanawha County, reflecting the area of more intensive sampling effort by the senior author. This imbalance of data between Kanawha County and other counties should even out with further sampling of the area. Key Words: Appalachian Mountains, -

Micro-Moth Grading Guidelines (Scotland) Abhnumber Code
Micro-moth Grading Guidelines (Scotland) Scottish Adult Mine Case ABHNumber Code Species Vernacular List Grade Grade Grade Comment 1.001 1 Micropterix tunbergella 1 1.002 2 Micropterix mansuetella Yes 1 1.003 3 Micropterix aureatella Yes 1 1.004 4 Micropterix aruncella Yes 2 1.005 5 Micropterix calthella Yes 2 2.001 6 Dyseriocrania subpurpurella Yes 2 A Confusion with fly mines 2.002 7 Paracrania chrysolepidella 3 A 2.003 8 Eriocrania unimaculella Yes 2 R Easier if larva present 2.004 9 Eriocrania sparrmannella Yes 2 A 2.005 10 Eriocrania salopiella Yes 2 R Easier if larva present 2.006 11 Eriocrania cicatricella Yes 4 R Easier if larva present 2.007 13 Eriocrania semipurpurella Yes 4 R Easier if larva present 2.008 12 Eriocrania sangii Yes 4 R Easier if larva present 4.001 118 Enteucha acetosae 0 A 4.002 116 Stigmella lapponica 0 L 4.003 117 Stigmella confusella 0 L 4.004 90 Stigmella tiliae 0 A 4.005 110 Stigmella betulicola 0 L 4.006 113 Stigmella sakhalinella 0 L 4.007 112 Stigmella luteella 0 L 4.008 114 Stigmella glutinosae 0 L Examination of larva essential 4.009 115 Stigmella alnetella 0 L Examination of larva essential 4.010 111 Stigmella microtheriella Yes 0 L 4.011 109 Stigmella prunetorum 0 L 4.012 102 Stigmella aceris 0 A 4.013 97 Stigmella malella Apple Pigmy 0 L 4.014 98 Stigmella catharticella 0 A 4.015 92 Stigmella anomalella Rose Leaf Miner 0 L 4.016 94 Stigmella spinosissimae 0 R 4.017 93 Stigmella centifoliella 0 R 4.018 80 Stigmella ulmivora 0 L Exit-hole must be shown or larval colour 4.019 95 Stigmella viscerella -

Molecular Basis of Pheromonogenesis Regulation in Moths
Chapter 8 Molecular Basis of Pheromonogenesis Regulation in Moths J. Joe Hull and Adrien Fónagy Abstract Sexual communication among the vast majority of moths typically involves the synthesis and release of species-specifc, multicomponent blends of sex pheromones (types of insect semiochemicals) by females. These compounds are then interpreted by conspecifc males as olfactory cues regarding female reproduc- tive readiness and assist in pinpointing the spatial location of emitting females. Studies by multiple groups using different model systems have shown that most sex pheromones are synthesized de novo from acetyl-CoA by functionally specialized cells that comprise the pheromone gland. Although signifcant progress was made in identifying pheromone components and elucidating their biosynthetic pathways, it wasn’t until the advent of modern molecular approaches and the increased avail- ability of genetic resources that a more complete understanding of the molecular basis underlying pheromonogenesis was developed. Pheromonogenesis is regulated by a neuropeptide termed Pheromone Biosynthesis Activating Neuropeptide (PBAN) that acts on a G protein-coupled receptor expressed at the surface of phero- mone gland cells. Activation of the PBAN receptor (PBANR) triggers a signal trans- duction cascade that utilizes an infux of extracellular Ca2+ to drive the concerted action of multiple enzymatic steps (i.e. chain-shortening, desaturation, and fatty acyl reduction) that generate the multicomponent pheromone blends specifc to each species. In this chapter, we provide a brief overview of moth sex pheromones before expanding on the molecular mechanisms regulating pheromonogenesis, and con- clude by highlighting recent developments in the literature that disrupt/exploit this critical pathway. J. J. Hull (*) USDA-ARS, US Arid Land Agricultural Research Center, Maricopa, AZ, USA e-mail: [email protected] A. -

Biodiversity Climate Change Impacts Report Card Technical Paper 12. the Impact of Climate Change on Biological Phenology In
Sparks Pheno logy Biodiversity Report Card paper 12 2015 Biodiversity Climate Change impacts report card technical paper 12. The impact of climate change on biological phenology in the UK Tim Sparks1 & Humphrey Crick2 1 Faculty of Engineering and Computing, Coventry University, Priory Street, Coventry, CV1 5FB 2 Natural England, Eastbrook, Shaftesbury Road, Cambridge, CB2 8DR Email: [email protected]; [email protected] 1 Sparks Pheno logy Biodiversity Report Card paper 12 2015 Executive summary Phenology can be described as the study of the timing of recurring natural events. The UK has a long history of phenological recording, particularly of first and last dates, but systematic national recording schemes are able to provide information on the distributions of events. The majority of data concern spring phenology, autumn phenology is relatively under-recorded. The UK is not usually water-limited in spring and therefore the major driver of the timing of life cycles (phenology) in the UK is temperature [H]. Phenological responses to temperature vary between species [H] but climate change remains the major driver of changed phenology [M]. For some species, other factors may also be important, such as soil biota, nutrients and daylength [M]. Wherever data is collected the majority of evidence suggests that spring events have advanced [H]. Thus, data show advances in the timing of bird spring migration [H], short distance migrants responding more than long-distance migrants [H], of egg laying in birds [H], in the flowering and leafing of plants[H] (although annual species may be more responsive than perennial species [L]), in the emergence dates of various invertebrates (butterflies [H], moths [M], aphids [H], dragonflies [M], hoverflies [L], carabid beetles [M]), in the migration [M] and breeding [M] of amphibians, in the fruiting of spring fungi [M], in freshwater fish migration [L] and spawning [L], in freshwater plankton [M], in the breeding activity among ruminant mammals [L] and the questing behaviour of ticks [L]. -

African Butterfly News Can Be Downloaded Here
LATE SUMMER EDITION: JANUARY / AFRICAN FEBRUARY 2018 - 1 BUTTERFLY THE LEPIDOPTERISTS’ SOCIETY OF AFRICA NEWS LATEST NEWS Welcome to the first newsletter of 2018! I trust you all have returned safely from your December break (assuming you had one!) and are getting into the swing of 2018? With few exceptions, 2017 was a very poor year butterfly-wise, at least in South Africa. The drought continues to have a very negative impact on our hobby, but here’s hoping that 2018 will be better! Braving the Great Karoo and Noorsveld (Mark Williams) In the first week of November 2017 Jeremy Dobson and I headed off south from Egoli, at the crack of dawn, for the ‘Harde Karoo’. (Is there a ‘Soft Karoo’?) We had a very flexible plan for the six-day trip, not even having booked any overnight accommodation. We figured that finding a place to commune with Uncle Morpheus every night would not be a problem because all the kids were at school. As it turned out we did not have to spend a night trying to kip in the Pajero – my snoring would have driven Jeremy nuts ... Friday 3 November The main purpose of the trip was to survey two quadrants for the Karoo BioGaps Project. One of these was on the farm Lushof, 10 km west of Loxton, and the other was Taaiboschkloof, about 50 km south-east of Loxton. The 1 000 km drive, via Kimberley, to Loxton was accompanied by hot and windy weather. The temperature hit 38 degrees and was 33 when the sun hit the horizon at 6 pm. -

Species List
Species List for <vice county> [Staffordshire (VC 39)] Code Taxon Vernacular 1.001 Micropterix tunbergella 1.002 Micropterix mansuetella 1.003 Micropterix aureatella 1.004 Micropterix aruncella 1.005 Micropterix calthella 2.001 Dyseriocrania subpurpurella 2.003 Eriocrania unimaculella 2.004 Eriocrania sparrmannella 2.005 Eriocrania salopiella 2.006 Eriocrania cicatricella 2.006 Eriocrania haworthi 2.007 Eriocrania semipurpurella 2.008 Eriocrania sangii 3.001 Triodia sylvina Orange Swift 3.002 Korscheltellus lupulina Common Swift 3.003 Korscheltellus fusconebulosa Map-winged Swift 3.004 Phymatopus hecta Gold Swift 3.005 Hepialus humuli Ghost Moth 4.002 Stigmella lapponica 4.003 Stigmella confusella 4.004 Stigmella tiliae 4.005 Stigmella betulicola 4.006 Stigmella sakhalinella 4.007 Stigmella luteella 4.008 Stigmella glutinosae 4.009 Stigmella alnetella 4.010 Stigmella microtheriella 4.012 Stigmella aceris 4.013 Stigmella malella Apple Pygmy 4.015 Stigmella anomalella Rose Leaf Miner 4.017 Stigmella centifoliella 4.018 Stigmella ulmivora 4.019 Stigmella viscerella 4.020 Stigmella paradoxa 4.022 Stigmella regiella 4.023 Stigmella crataegella 4.024 Stigmella magdalenae 4.025 Stigmella nylandriella 4.026 Stigmella oxyacanthella 4.030 Stigmella hybnerella 4.032 Stigmella floslactella 4.034 Stigmella tityrella 4.035 Stigmella salicis 4.036 Stigmella myrtillella 4.038 Stigmella obliquella 4.039 Stigmella trimaculella 4.040 Stigmella assimilella 4.041 Stigmella sorbi 4.042 Stigmella plagicolella 4.043 Stigmella lemniscella 4.044 Stigmella continuella -

Butterflies and Moths of Snohomish County, Washington, United
Heliothis ononis Flax Bollworm Moth Coptotriche aenea Blackberry Leafminer Argyresthia canadensis Apyrrothrix araxes Dull Firetip Phocides pigmalion Mangrove Skipper Phocides belus Belus Skipper Phocides palemon Guava Skipper Phocides urania Urania skipper Proteides mercurius Mercurial Skipper Epargyreus zestos Zestos Skipper Epargyreus clarus Silver-spotted Skipper Epargyreus spanna Hispaniolan Silverdrop Epargyreus exadeus Broken Silverdrop Polygonus leo Hammock Skipper Polygonus savigny Manuel's Skipper Chioides albofasciatus White-striped Longtail Chioides zilpa Zilpa Longtail Chioides ixion Hispaniolan Longtail Aguna asander Gold-spotted Aguna Aguna claxon Emerald Aguna Aguna metophis Tailed Aguna Typhedanus undulatus Mottled Longtail Typhedanus ampyx Gold-tufted Skipper Polythrix octomaculata Eight-spotted Longtail Polythrix mexicanus Mexican Longtail Polythrix asine Asine Longtail Polythrix caunus (Herrich-Schäffer, 1869) Zestusa dorus Short-tailed Skipper Codatractus carlos Carlos' Mottled-Skipper Codatractus alcaeus White-crescent Longtail Codatractus yucatanus Yucatan Mottled-Skipper Codatractus arizonensis Arizona Skipper Codatractus valeriana Valeriana Skipper Urbanus proteus Long-tailed Skipper Urbanus viterboana Bluish Longtail Urbanus belli Double-striped Longtail Urbanus pronus Pronus Longtail Urbanus esmeraldus Esmeralda Longtail Urbanus evona Turquoise Longtail Urbanus dorantes Dorantes Longtail Urbanus teleus Teleus Longtail Urbanus tanna Tanna Longtail Urbanus simplicius Plain Longtail Urbanus procne Brown Longtail -
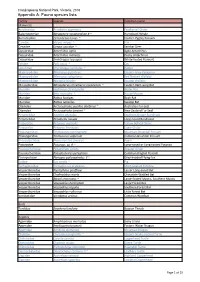
Report-VIC-Croajingolong National Park-Appendix A
Croajingolong National Park, Victoria, 2016 Appendix A: Fauna species lists Family Species Common name Mammals Acrobatidae Acrobates pygmaeus Feathertail Glider Balaenopteriae Megaptera novaeangliae # ~ Humpback Whale Burramyidae Cercartetus nanus ~ Eastern Pygmy Possum Canidae Vulpes vulpes ^ Fox Cervidae Cervus unicolor ^ Sambar Deer Dasyuridae Antechinus agilis Agile Antechinus Dasyuridae Antechinus mimetes Dusky Antechinus Dasyuridae Sminthopsis leucopus White-footed Dunnart Felidae Felis catus ^ Cat Leporidae Oryctolagus cuniculus ^ Rabbit Macropodidae Macropus giganteus Eastern Grey Kangaroo Macropodidae Macropus rufogriseus Red Necked Wallaby Macropodidae Wallabia bicolor Swamp Wallaby Miniopteridae Miniopterus schreibersii oceanensis ~ Eastern Bent-wing Bat Muridae Hydromys chrysogaster Water Rat Muridae Mus musculus ^ House Mouse Muridae Rattus fuscipes Bush Rat Muridae Rattus lutreolus Swamp Rat Otariidae Arctocephalus pusillus doriferus ~ Australian Fur-seal Otariidae Arctocephalus forsteri ~ New Zealand Fur Seal Peramelidae Isoodon obesulus Southern Brown Bandicoot Peramelidae Perameles nasuta Long-nosed Bandicoot Petauridae Petaurus australis Yellow Bellied Glider Petauridae Petaurus breviceps Sugar Glider Phalangeridae Trichosurus cunninghami Mountain Brushtail Possum Phalangeridae Trichosurus vulpecula Common Brushtail Possum Phascolarctidae Phascolarctos cinereus Koala Potoroidae Potorous sp. # ~ Long-nosed or Long-footed Potoroo Pseudocheiridae Petauroides volans Greater Glider Pseudocheiridae Pseudocheirus peregrinus