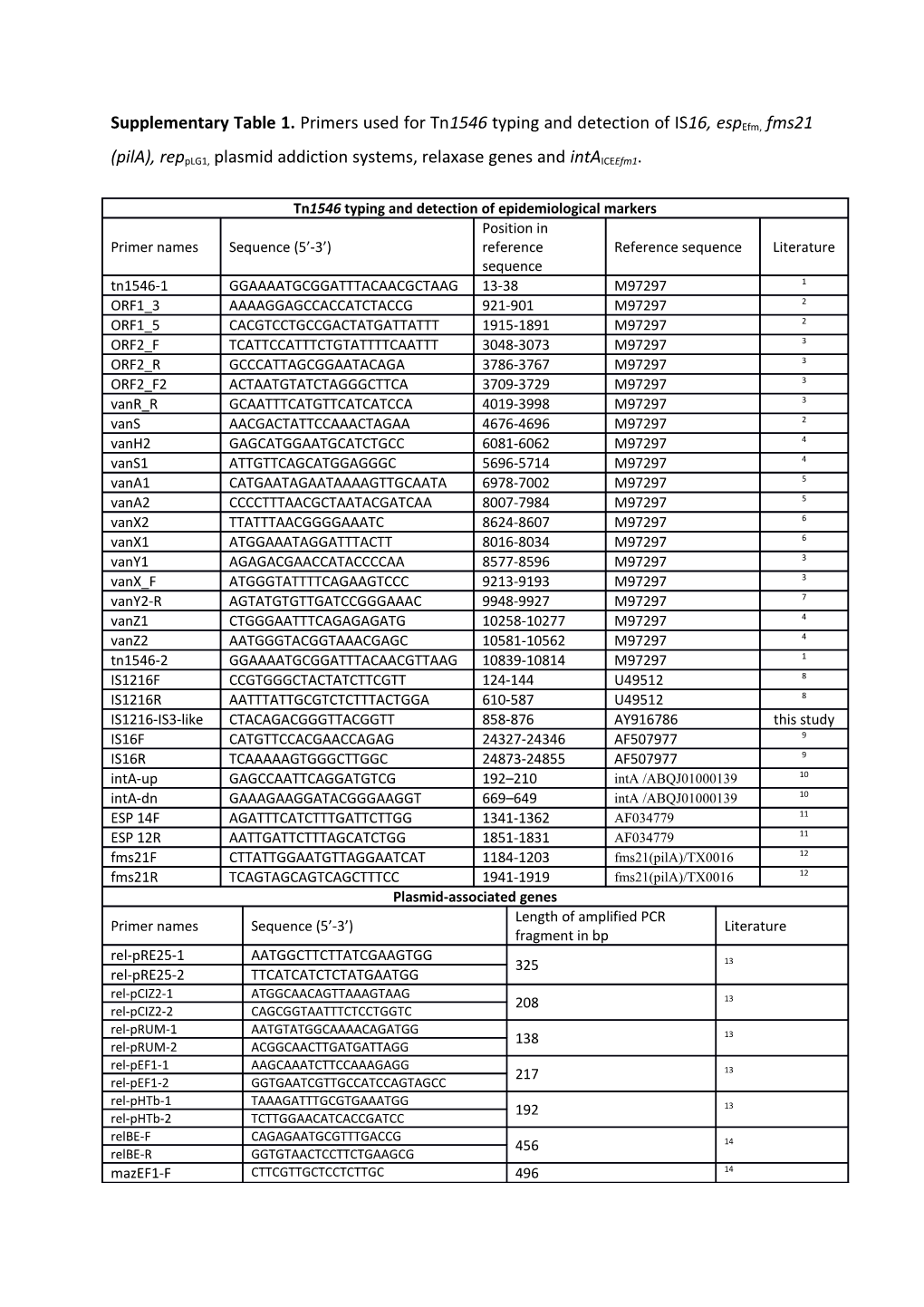
Supplementary Table 1. Primers used for Tn1546 typing and detection of IS16, espEfm, fms21
(pilA), reppLG1, plasmid addiction systems, relaxase genes and intAICEEfm1.
Tn1546 typing and detection of epidemiological markers Position in Primer names Sequence (5’-3’) reference Reference sequence Literature sequence tn1546-1 GGAAAATGCGGATTTACAACGCTAAG 13-38 M97297 1 ORF1_3 AAAAGGAGCCACCATCTACCG 921-901 M97297 2 ORF1_5 CACGTCCTGCCGACTATGATTATTT 1915-1891 M97297 2 ORF2_F TCATTCCATTTCTGTATTTTCAATTT 3048-3073 M97297 3 ORF2_R GCCCATTAGCGGAATACAGA 3786-3767 M97297 3 ORF2_F2 ACTAATGTATCTAGGGCTTCA 3709-3729 M97297 3 vanR_R GCAATTTCATGTTCATCATCCA 4019-3998 M97297 3 vanS AACGACTATTCCAAACTAGAA 4676-4696 M97297 2 vanH2 GAGCATGGAATGCATCTGCC 6081-6062 M97297 4 vanS1 ATTGTTCAGCATGGAGGGC 5696-5714 M97297 4 vanA1 CATGAATAGAATAAAAGTTGCAATA 6978-7002 M97297 5 vanA2 CCCCTTTAACGCTAATACGATCAA 8007-7984 M97297 5 vanX2 TTATTTAACGGGGAAATC 8624-8607 M97297 6 vanX1 ATGGAAATAGGATTTACTT 8016-8034 M97297 6 vanY1 AGAGACGAACCATACCCCAA 8577-8596 M97297 3 vanX_F ATGGGTATTTTCAGAAGTCCC 9213-9193 M97297 3 vanY2-R AGTATGTGTTGATCCGGGAAAC 9948-9927 M97297 7 vanZ1 CTGGGAATTTCAGAGAGATG 10258-10277 M97297 4 vanZ2 AATGGGTACGGTAAACGAGC 10581-10562 M97297 4 tn1546-2 GGAAAATGCGGATTTACAACGTTAAG 10839-10814 M97297 1 IS1216F CCGTGGGCTACTATCTTCGTT 124-144 U49512 8 IS1216R AATTTATTGCGTCTCTTTACTGGA 610-587 U49512 8 IS1216-IS3-like CTACAGACGGGTTACGGTT 858-876 AY916786 this study IS16F CATGTTCCACGAACCAGAG 24327-24346 AF507977 9 IS16R TCAAAAAGTGGGCTTGGC 24873-24855 AF507977 9 intA-up GAGCCAATTCAGGATGTCG 192–210 intA /ABQJ01000139 10 intA-dn GAAAGAAGGATACGGGAAGGT 669–649 intA /ABQJ01000139 10 ESP 14F AGATTTCATCTTTGATTCTTGG 1341-1362 AF034779 11 ESP 12R AATTGATTCTTTAGCATCTGG 1851-1831 AF034779 11 fms21F CTTATTGGAATGTTAGGAATCAT 1184-1203 fms21(pilA)/TX0016 12 fms21R TCAGTAGCAGTCAGCTTTCC 1941-1919 fms21(pilA)/TX0016 12 Plasmid-associated genes Length of amplified PCR Primer names Sequence (5’-3’) Literature fragment in bp rel-pRE25-1 AATGGCTTCTTATCGAAGTGG 325 13 rel-pRE25-2 TTCATCATCTCTATGAATGG rel-pCIZ2-1 ATGGCAACAGTTAAAGTAAG 208 13 rel-pCIZ2-2 CAGCGGTAATTTCTCCTGGTC rel-pRUM-1 AATGTATGGCAAAACAGATGG 138 13 rel-pRUM-2 ACGGCAACTTGATGATTAGG rel-pEF1-1 AAGCAAATCTTCCAAAGAGG 217 13 rel-pEF1-2 GGTGAATCGTTGCCATCCAGTAGCC rel-pHTb-1 TAAAGATTTGCGTGAAATGG 192 13 rel-pHTb-2 TCTTGGAACATCACCGATCC relBE-F CAGAGAATGCGTTTGACCG 456 14 relBE-R GGTGTAACTCCTTCTGAAGCG mazEF1-F CTTCGTTGCTCCTCTTGC 496 14 mazEF1-R CGTTGGGGAAATTCACCG axe-txe-F CTGACCCTTTCCTTACTTCCG 556 14 axe-txe-R GGGTGAAAGGAATGGAAGCAG e-z-F GTGGTTTAGGTGGCTGCAAG 1044 14 e-z-R TTAACGAATTATCGGCAAGC efm-repA_pLG1-up GAAAATGATATCTACTTACTCG 568 7 efm-repA_pLG1-dn TTACATAGACAAAAATCAGGT
1. Palepou MF, Adebiyi AM, Tremlett CH et al. Molecular analysis of diverse elements mediating VanA glycopeptide resistance in enterococci. J Antimicrob Chemother 1998; 42: 605-12. 2. Huh JY, Lee WG, Lee K et al. Distribution of insertion sequences associated with Tn1546-Like elements among Enterococcus faecium isolates from patients in Korea. J Clin Microbiol 2004; 42: 1897-902. 3. Talebi M, Pourshafie MR, Katouli M et al. Molecular structure and transferability of Tn1546- like elements in Enterococcus faecium isolates from clinical, sewage, and surface water samples in Iran. Appl Environ Microbiol 2008; 74: 1350-6. 4. Jensen LB, Ahrens P, Dons L et al. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J Clin Microbiol 1998; 36: 437-42. 5. Clark NC, Cooksey RC, Hill BC et al. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother 1993; 37: 2311-7. 6. Yu HS, Seol SY, Cho DT. Diversity of Tn1546-like elements in vancomycin-resistant enterococci isolated from humans and poultry in Korea. J Clin Microbiol 2003; 41: 2641-3. 7. Wardal E, Markowska K, Zabicka D et al. Molecular analysis of VanA outbreak of Enterococcus faecium in two Warsaw hospitals: the importance of mobile genetic elements. Biomed Res Int 2014: 575367. 8. Tsai JC, Hsueh PR, Chen HJ et al. The erm(T) gene is flanked by IS1216V in inducible erythromycin-resistant Streptococcus gallolyticus subsp. pasteurianus. Antimicrob Agents Chemother 2005; 49: 4347-50. 9. Werner G, Fleige C, Geringer U et al. IS element IS16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect Dis 2011; 11: 80. 10. Sadowy E, Sienko A, Gawryszewska I et al. High abundance and diversity of antimicrobial resistance determinants among early vancomycin-resistant Enterococcus faecium in Poland. Eur J Clin Microbiol Infect Dis 2013; 32: 1193-203. 11. Vankerckhoven V, Van Autgaerden T, Vael C et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol 2004; 42: 4473-9. 12. Sillanpaa J, Nallapareddy SR, Prakash VP et al. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology 2008; 154: 3199-211. 13. Freitas AR. Ecology and evolution of antimicrobial resistance in Enterococcus: A multilayered molecular approach with emphasis in plasmid diversity. PhD Thesis 2011. 14. Moritz EM, Hergenrother PJ. Toxin-antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc Natl Acad Sci U S A 2007; 104: 311-6.