1 Supplemental Tables
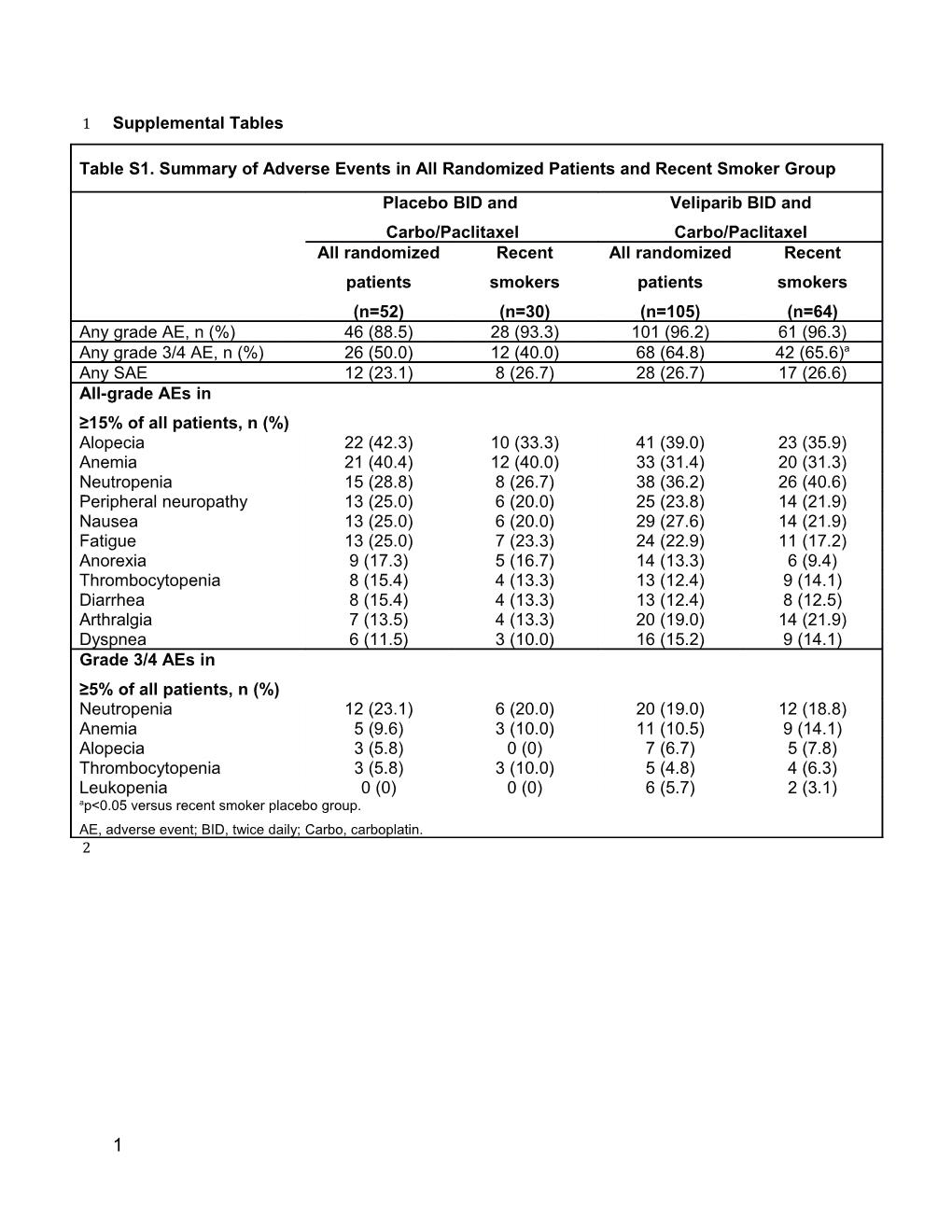
Table S1. Summary of Adverse Events in All Randomized Patients and Recent Smoker Group Placebo BID and Veliparib BID and Carbo/Paclitaxel Carbo/Paclitaxel All randomized Recent All randomized Recent patients smokers patients smokers (n=52) (n=30) (n=105) (n=64) Any grade AE, n (%) 46 (88.5) 28 (93.3) 101 (96.2) 61 (96.3) Any grade 3/4 AE, n (%) 26 (50.0) 12 (40.0) 68 (64.8) 42 (65.6)a Any SAE 12 (23.1) 8 (26.7) 28 (26.7) 17 (26.6) All-grade AEs in ≥15% of all patients, n (%) Alopecia 22 (42.3) 10 (33.3) 41 (39.0) 23 (35.9) Anemia 21 (40.4) 12 (40.0) 33 (31.4) 20 (31.3) Neutropenia 15 (28.8) 8 (26.7) 38 (36.2) 26 (40.6) Peripheral neuropathy 13 (25.0) 6 (20.0) 25 (23.8) 14 (21.9) Nausea 13 (25.0) 6 (20.0) 29 (27.6) 14 (21.9) Fatigue 13 (25.0) 7 (23.3) 24 (22.9) 11 (17.2) Anorexia 9 (17.3) 5 (16.7) 14 (13.3) 6 (9.4) Thrombocytopenia 8 (15.4) 4 (13.3) 13 (12.4) 9 (14.1) Diarrhea 8 (15.4) 4 (13.3) 13 (12.4) 8 (12.5) Arthralgia 7 (13.5) 4 (13.3) 20 (19.0) 14 (21.9) Dyspnea 6 (11.5) 3 (10.0) 16 (15.2) 9 (14.1) Grade 3/4 AEs in ≥5% of all patients, n (%) Neutropenia 12 (23.1) 6 (20.0) 20 (19.0) 12 (18.8) Anemia 5 (9.6) 3 (10.0) 11 (10.5) 9 (14.1) Alopecia 3 (5.8) 0 (0) 7 (6.7) 5 (7.8) Thrombocytopenia 3 (5.8) 3 (10.0) 5 (4.8) 4 (6.3) Leukopenia 0 (0) 0 (0) 6 (5.7) 2 (3.1) ap<0.05 versus recent smoker placebo group. AE, adverse event; BID, twice daily; Carbo, carboplatin. 2
1 3
Table S2. Paclitaxel Plasma Concentrations (ng/mL) by Cotinine Level Patients with Cotinine ≤10 ng/mL Patients with Cotinine >10 ng/mL C1D3 C2D3 C3D3 C4D3 C1D3 C2D3 C3D3 C4D3 Placebo BID N=29 N=29 N=24 N=21 N=21 N=18 N=15 N=13 6133 ± 5868 ± 7185 ± 7001 ± 4182 ± 9227 ± 5810 ± 7992 ± 2700 2345 5035 3754 2499 12482 4596 9530 (5960) (5810) (6290) (6570) (4300) (6380) (4760) (5780) Veliparib 120 mg BID N=58 N=46 N=38 N=35 N=40 N=37 N=32 N=32 12842 ± 5926 ± 5824 ± 8809 ± 13441 ± 10041 ± 6584 ± 6798 ± 39061 3024 2656 16544 41487 22330 5693 3198 (5565) (5310) (5635) (5930) (6680) (6350) (5615) (5865) Data are mean ± SD (median) plasma concentrations of paclitaxel just before ending paclitaxel infusion (2 hr 55 min after start of paclitaxel infusion) in patients with cotinine concentrations ≤10 ng/mL and >10 ng/mL at screening. BID, twice daily; C, cycle; D, day; SD, standard deviation. 4 5
2 Table S3. Carboplatin Plasma Concentrations (µg/mL) by Cotinine Level Patients with Cotinine ≤10 ng/mL Patients with Cotinine >10 ng/mL C1D3 C2D3 C3D3 C4D3 C1D3 C2D3 C3D3 C4D3 Placebo BID N=29 N=28 N=24 N=21 N=21 N=18 N=15 N=13 29.6 ± 33.5 ± 56.2 ± 37.8 ± 30.7 ± 29.1 ± 13 30.6 ± 39.9 ± 18.3 21.8 103.3 16.4 14.3 (30.9) 14.3 17.9 (31.6) (38.5) (35.2) (31.8) (35.2) (37.9) (35.0) Veliparib 120 mg BID N=58 N=46 N=39 N=36 N=40 N=39 N=33 N=34 31.2 ± 31.3 ± 27.9 ± 42.6 ± 33.6 ± 38.0 ± 34.9 ± 34.5 ± 14.1 18.5 15.3 46.4 15.6 42.9 16.2 15.0 (32.7) (33.0) (32) (35.1) (30.7) (33.3) (35.5) (32.4) Data are mean ± SD (median) plasma concentrations of carboplatin just before ending carboplatin infusion (25 min after start) in patients with cotinine concentrations ≤10 ng/mL and >10 ng/mL at screening. BID, twice daily; C, cycle; D, day; SD, standard deviation. 6 7 8
3 9 Table S4. Summary of Primary and Secondary Endpoints by Smoking History Placebo BID and Veliparib BID and HR Adjusted HRa Carbo/Paclitaxel Carbo/Paclitaxel Median PFS, months (95% CI) 4.2 (3.1–5.6) 5.8 (4.2–6.1) 0.74 (0.46–1.17) 0.57 (0.35–0.92) Recent 3.3 (1.4–4.2) 5.6 (4.1–7.0) 0.38 (0.21–0.67) 0.37 (0.21–0.68) Former NA (3.3–NA) 6.0 (2.4–NA) 2.10 (0.66–6.65) 0.77 (0.20–3.06) Never 5.6 (1.4–8.2) 6.4 (1.0–NA) 1.03 (0.27–3.85) 0.96 (0.21–4.46) Median OS, months (95% CI) 9.1 (5.4–12.3) 11.7 (8.8–13.7) 0.80 (0.54–1.18) 0.72 (0.49–1.07) Recent 5.4 (3.8–8.8) 12.5 (9.9–16.6) 0.43 (0.26–0.70) 0.45 (0.27–0.76) Former 14.6 (9.2–NA) 8.6 (5.9–17.5) 1.62 (0.73–3.6) 0.72 (0.27–1.92) Never NA (3.6–NA) 13.2 (5.0–NA) 1.34 (0.40–4.44) 0.71 (0.18–2.74) ORR, % (95% CI) 32 (20–46) 32 (24–42) ------Recent 26 (12–45) 31 (20–44) Former 43 (18–71) 39 (22–59) Never 38 (9–76) 23 (5–54) Median DOR, months (95% CI) 3.3 (2.7–5.3) 6.9 (4.4–7.0) 0.11 (0.03–0.50) ----- Recent 4.3 (1.5–NA) 6.9 (4.4–7.0) 0.39 (0.09–1.66) Former NA (2.9–NA) 5.5 (4.6–NA) 0.51 (0.07–3.67) Never NA (2.8–NA) NA (5.0–NA) 0.51 (0.07–3.67) aFrom Cox proportional hazard model, adjusted for gender and baseline ECOG PS. BID, twice daily; Carbo, carboplatin; CI, confidence interval; DOR, duration of overall response; ECOG PS, Eastern Cooperative Oncology Group performance status; HR, hazard ratio; NA, not available; ORR, objective response rate; OS, overall survival; PFS, progression-free survival. 10 11
4 12 Table S5. Impact of Smoking During Study Treatment in Recent Smokers
Placebo BID and Veliparib BID and Carbo/Paclitaxel Carbo/Paclitaxel HR Cotinine high: n=22 Cotinine high: n=41 Cotinine low: n=8 Cotinine low: n=23
Median PFS, months (95% CI)
Cotinine-high 3.5 (1.3–5.6) 5.6 (4.3–5.9) 0.38 (0.19–0.75) recent smokers
Cotinine-low 3.1 (0.0–5.0) 5.8 (2.5–8.4) 0.43 (0.15–1.23) recent smokers
Median OS, months (95% CI)
Cotinine-high 5.7 (3.6–11.1) 12.5 (9.5–17.0) 0.50 (0.27–0.90) recent smokers
Cotinine-low recent smokers 4.9 (0.0–8.4) 12.8 (7.9–17.7) 0.22 (0.08–0.61)
BID, twice daily; Carbo, carboplatin; CI, confidence interval; HR, hazard ratio; OS, overall survival; PFS, progression-free survival. 13
5 14
Table S6. Hazard Ratios in Smoking Status Subgroup Based on a Multivariate Cox Proportional Hazards Model for Overall Survival Endpoint
From Multivariate Modela HR 95% CI of HR
V vs P in current smokers 0.4541 0.2157–0.9560
V vs P in past smokers 1.4416 0.5916–3.5125
V vs P in never smokers 0.9663 0.2421–3.8562
aModel includes treatment, smoking history, histology, age, ECOG, gender, geographic region. CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio; P, placebo; V, veliparib. 15 16
6 17
Table S7. Hazard Ratios in Smoking Status Subgroup Based on a Multivariate Cox PH Model for Progression Free Survival Endpoint
From Multivariate Model** HR 95% CI of HR
V vs P in Current smokers 0.4090 0.1744–0.9593
V vs P in Past Smokers 1.3352 0.3755–4.7482
V vs P in never smoked 0.7245 0.1535–3.4192
**Model includes treatment, smoking status, Histology, age group, ECOG, gender, Region. CI, confidence interval; HR, hazard ratio; P, placebo; V, veliparib. 18 19
7 20 21 Supplemental Figures 22
23 Figure S1. Case Report Form Example 24
25
8 26
9 27 Supplemental Methods
28 DNA Extraction, Exome Sequencing, and Variant Calling
29
30 Genomic DNA (and RNA) was extracted using Qiagen AllPrep kit. Profiling aimed to achieve a
31 150X mean on-target coverage for tumor (and 100X for germline DNA). Alignment was
32 performed using OSA (http://omicsoft.com/downloads/whitepaper/OmicsoftAligner.pd). Somatic
33 call was performed by Array Studio module “Summarize Matched Pair Variation Data” (the
34 same algorithm as Varscan2) to identify somatic mutations. A one-sided Fisher exact test is
35 done for the somatic mutation test. For variants called by ArrayStudio, filters include: Base
36 Quality Cutoff is 13, Map Quality Cutoff is 0, Minimal coverage in normal samples is 8, Minimal
37 coverage in Tumor samples is 6, Minimal Mutation Hit is 2, Frequency Difference Cutoff is 0.20,
38 Filtering Significance Level=0.05, Maximal Frequency Cutoff is 0.10. In order to remove false
39 positive calls due to sequencing or alignment-related artifacts, a set of filters were set up
40 (Koboldt D.K. et al, Genome Research, 2012). These include: Read Position Cutoff is 0.10,
41 Strandness Cutoff is 0.9, Homopolymer Cutoff is 5, Mapping Quality Difference Cutoff is 30,
42 Read Length Difference Cutoff is 25, Mmqs Difference (Difference in average mismatch quality
43 sum between variant and reference reads) Cutoff is 100. There are two categories in the variant
44 calls –high confidence or low confidence, only high confidence somatic calls were kept to make
45 somatic mutational signature and mutational burden calculation. Variants in protein coding
46 regions and splicing sites were used to calculate mutational signature (Alexandrov L.B. et al.,
47 Nature, 2013). These are the so called “tier 1” mutations in whole exome sequencing data
48 analysis (Mardis E.R. et al, N Engl J Med. 2009;361). Certain germline variants still leaked
49 through after somatic calling. Common SNPs (defined as variants with population frequency =
50 or >1% in dbSNP database or ExAC database) were further filtered out. When calculating
51 somatic mutational signature, functional significance of the variants were not evaluated as even
52 silent mutations (or neutral mutations) could be informative for the underlying mutagenesis
10 53 processes (Alexandrov L.B. et al., Nature 2013; Helleday T. et al., Nat Rev Genet 2014;
54 Alexandrov L.B. and Stratton M.R., Curr Opin Genet Dev 2014). Since only high quality exome
55 pairs (defined as of more than 90% of target exome region being sequenced more than 10
56 times) were used for our analysis and exomic region coverage is comparable across these high
57 quality exome pairs, no further normalization was done when comparing mutational burden.
58
59 Determination of Molecular Signatures
60
61 The deconvolution method as described in Rosenthal et al, 2016 was followed. To validate the
62 methodology, we downloaded TCGA ovarian and lung cancer somatic mutational data,
63 calculated somatic mutational signatures for each patient, and re-established the reported
64 correlation between age of onset and signature 1, smoking exposure and signature 4, and
65 BRCA1/2 mutational status and signature 3 (Alexandrov L.B. et al., Nature 2013; Helleday T. et
66 al., Nat Rev Genet 2014; Alexandrov L.B. and Stratton M.R., Curr Opin Genet Dev 2014).
67 This deconvolution method returned a numeric value for each somatic signature in each patient
68 tumor. We define scores of “zero” as “negative”, any value “more than zero” as “positive”.
69
11 70 List of supplemental materials 71
72 Supplemental Table 1. Word document file. 73 Supplemental Table 2. Word document file. 74 Supplemental Table 3. Word document file. 75 Supplemental Table 4. Word document file. 76 Supplemental Table 5. Word document file. 77 Supplemental Figure 1. Word documents file. 78 Supplemental Methods. Word document file. 79
80
12