1 Additional File 1
2
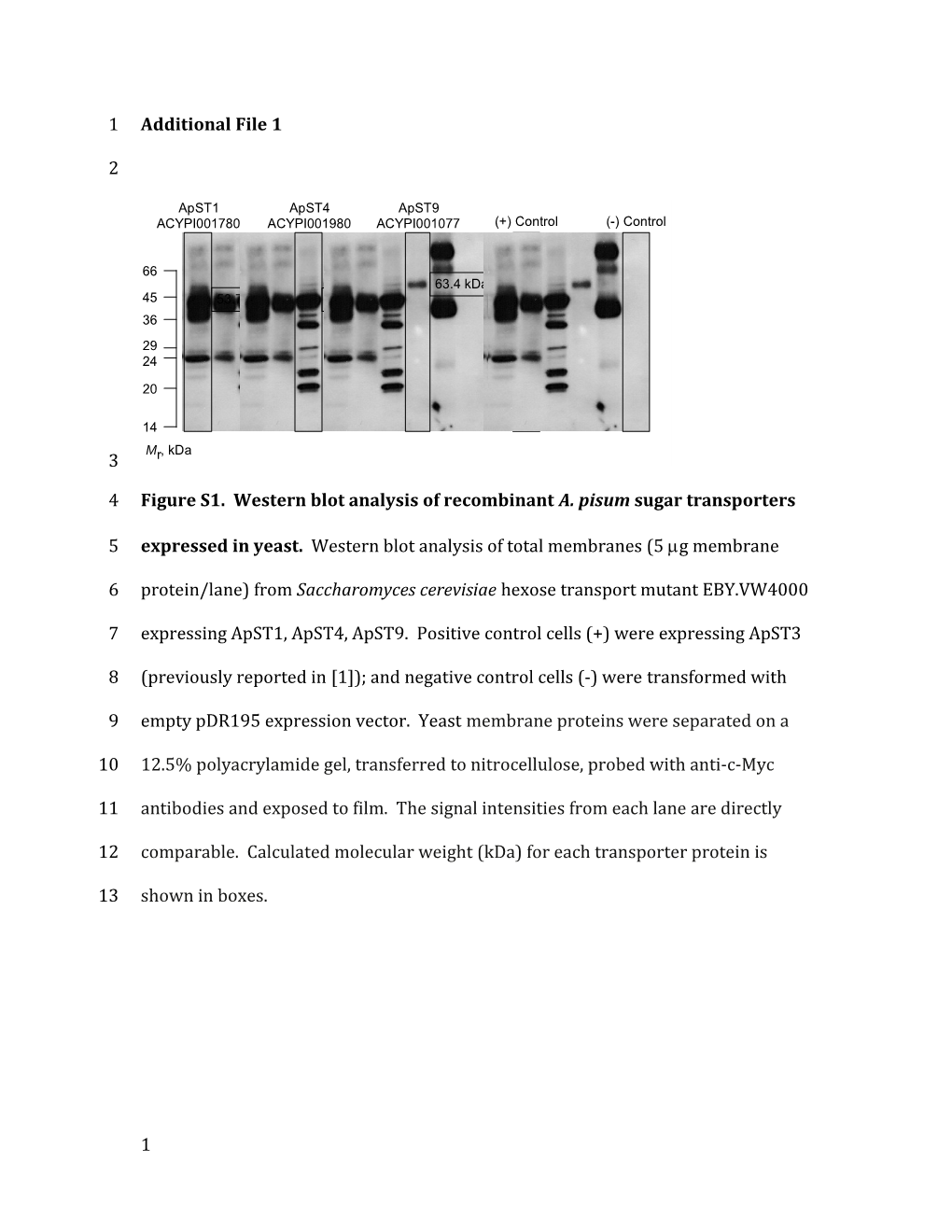
ApST1 ApST4 ApST9 ACYPI001780 ACYPI001980 ACYPI001077 (+) Control (-) Control
66 63.4 kDa 45 53.7 kDa 53.4 kDa 57.6 kDa 36 29 24
20
14 M , kDa 3 r
4 Figure S1. Western blot analysis of recombinant A. pisum sugar transporters
5 expressed in yeast. Western blot analysis of total membranes (5 g membrane
6 protein/lane) from Saccharomyces cerevisiae hexose transport mutant EBY.VW4000
7 expressing ApST1, ApST4, ApST9. Positive control cells (+) were expressing ApST3
8 (previously reported in [1]); and negative control cells (-) were transformed with
9 empty pDR195 expression vector. Yeast membrane proteins were separated on a
10 12.5% polyacrylamide gel, transferred to nitrocellulose, probed with anti-c-Myc
11 antibodies and exposed to film. The signal intensities from each lane are directly
12 comparable. Calculated molecular weight (kDa) for each transporter protein is
13 shown in boxes.
1 Helix 1 HuGLUT1 ------MEPSSKKLTGRLMLAVGGAVLG-SLQFGYNTGVINAPQKVIEEFYNQTW----VHRYG------53 HuGLUT4 ------MPSGFQQIGSEDGEPPQQRVTGTLVLAVFSAVLG-SLQFGYNIGVINAPQKVIEQSYNETW----LGRQGP------66 HuGLUT3 ------MGTQKVTPALIFAITVATIG-SFQFGYNTGVINAPEKIIKEFINKTL----TDKGN------51 HuGLUT2 ------MTEDKVTGTLVFTVITAVLG-SFQFGYDIGVINAPQQVIISHYRHVLGVPLDDRKAINNYVINSTDELPTISYSMNPKPTPW 81 ACYPI005975 MVAIKNELDTVQQVKEEIEELREYVDRLELQSHRRKLRLLEQGLTFFLSYTILASMLG-MLQFGYNTGVINAPEGNIEKFIKDVF----EDRYK------89 DmGLUT1 ------MAFLCAPGLTFFLTYSIFSAVLG-MLQFGYNTGVINAPEKNIENFMKDVY----KDRYG------54 ACYPI001980 ------MTEKQMVKDAEKQAPTPQNQSLNGRLLFAIIASAFGSAFQHGYNTGVVNAPQALIEKWISGVI----SGRNDGKP------71 AB550003 ------MSKEGEDIDEHQTKEELTKQNGVSGPDHATKGLNPRLAFAIAAAAVGSAFQHGYNLGVVNAPQKLIEEWILGVI----KNQTDASPP------83 AB549996 ------MNPDFSINLVFACMVAAIGAGFQHGYNTGVVNAPQNVIEKWMSDVS----QERHGMPP------54 .. * : : .* :*.**: **:***: * . . : Helix 2 Helix 3 Helix 4 Helix 5
HuGLUT1 ----ESILPTTLTTLWSLSVAIFSVGGMIGSFSVGLFVNRFGRRNSMLMMNLLAFVSAVLMGFSKLGKSFEMLILGRFIIGVYCGLTTGFVPMYVGEVSPTAFRGALGTLH QLGIVVGIL 169 HuGLUT4 -EGPSSIPPGTLTTLWALSVAIFSVGGMISSFLIGIISQWLGRKRAMLVNNVLAVLGGSLMGLANAAASYEMLILGRFLIGAYSGLTSGLVPMYVGEIAPTHLRGALGTLN QLAIVIGIL 185 HuGLUT3 ----APPSEVLLTSLWSLSVAIFSVGGMIGSFSVGLFVNRFGRRNSMLIVNLLAVTGGCFMGLCKVAKSVEMLILGRLVIGLFCGLCTGFVPMYIGEISPTALRGAFGTLN QLGIVVGIL 167 HuGLUT2 AEEETVAAAQLITMLWSLSVSSFAVGGMTASFFGGWLGDTLGRIKAMLVANILSLVGALLMGFSKLGPSHILIIAGRSISGLYCGLISGLVPMYIGEIAPTALRGALGTFH QLAIVTGIL 201 ACYPI005975 ----ENMDHGQAELLYSFAVSIFAIGGMLGGFSGGIIANRFGRKGGLLLNSFVGIGGACLMGLTKYFNSYEVLFIGRFIIGVNCGLNTSLVPMYISEIAPLNLRGGLGTVN QLAVTTGLL 205 DmGLUT1 ----EDISEEFIQQLYSVAVSIFAIGGMLGGFSGGWMANRFGRKGGLLLNNVLGIAGACLMGFTKVSHSYEMLFLGRFIIGVNCGLNTSLVPMYISEIAPLNLRGGLGTVN QLAVTVGLL 170 ACYPI001980 ------TDQTQVTLIWAVAVSIFCVGGMVGGLSTGFVAEKFGRKGGLLVSNALVILSAALQGVSKMYSSYELIIIGRFIIGINSGLNAGLTPMYLAEISPMNLRGSVGTVY QLVVTISIL 185 AB550003 ----SDANQTKVTMIFSIAVSIYCVGGMLGGAITGLVAEKYGRKGGLLFNNIFIVIAAALLGFSKAMNSYYMIIVGRFLLGINSGLNAGLTPMYLSEIAPVQLRGAVGTVY QLVLTISIL 199 AB549996 ------DKNDITFLFSLAVSIYCAGGIVGGLLTSTFAIHIGRRGGLFVNNLFALIAAAMMGLSKMAGSFELLIAGRCFSGLNSGLNSGLAGMYLVEVSPRSMRGALGSMY QLIITISIL 167 :::.:*: :. **: .. . . ** .::...... : *. : * ::: ** . * .** :.:. **: *::* :**..*:. ** :. .:* Helix 5 Helix 6 Helix 7
HuGLUT1 IAQVFGLDSIMGNKDLWPLLLSIIFIPALLQCIVLPFCPESPRFLLINRNEENRAKSVLKKLRGTADVTHDLQEMKEESRQMMREKKVTILELFRSPAYRQPILIAVVL QLSQQLSGINA 289 HuGLUT4 IAQVLGLESLLGTASLWPLLLGLTVLPALLQLVLLPFCPESPRYLYIIQNLEGPARKSLKRLTGWADVSGVLAELKDEKRKLERERPLSLLQLLGSRTHRQPLIIAVVL QLSQQLSGINA 305 HuGLUT3 VAQIFGLEFILGSEELWPLLLGFTILPAILQSAALPFCPESPRFLLINRKEEENAKQILQRLWGTQDVSQDIQEMKDESARMSQEKQVTVLELFRVSSYRQPIIISIVL QLSQQLSGINA 287 HuGLUT2 ISQIIGLEFILGNYDLWHILLGLSGVRAILQSLLLFFCPESPRYLYIKLDEEVKAKQSLKRLRGYDDVTKDINEMRKEREEASSEQKVSIIQLFTNSSYRQPILVALML HVAQQFSGING 321 ACYPI005975 ISQILGIEQILGTDEGWPLLLGLAICPAILQLILLPVCPESPRYLLITKQWEEEARKALRRLRATNQIEEDIEEMRAEERAQQSEATISMMELVCSPTLRQPLIISVVM QLSQQLSGINA 325 DmGLUT1 LSQVLGIEQILGTNEGWPILLGLAICPAILQLILLPVCPESPRYLLITKQWEEEARKALRRLRASGSVEEDIEEMRAEERAQQSESHISTMELICSPTLRPPLIIGIVM QLSQQFSGINA 290 ACYPI001980 ISQILGLDYILGTAELWPVLLALIIAPAIFMFATLPFCPESPKYTLINKKKDIEAERGLQWLRGTIEVHDEMDEMRAENEAMKVIPKVTLREMLSNPMLKTPLGISVMI MLCQQLSGINA 305 AB550003 ISQILGLNFILGTAELWPILLSLTIVPTIFQLITLPMCPESPKYLLITKGQEIESQRAVTWFRGTIEVHDEMDEMRREYESMKLVPKVTLREMLVNSALRIPLFISLVV MIAQQLSGINA 319 AB549996 VSQILGSQSIFGTDDLWPVLFGLTGIMALAQMLFLPCCPETPKH-IFNKGNKERAQKSLKWLRKREDVSAEMSEIQTEAEQEKSIGKASFQQFIQNPSLRKPLIIAIVI MIAQQLSGINA 286 ::*::* : ::*. . * :*:.: :: * ***:*:. : . :. : : .: : *:: * : ::. : *: :.::: :.**:****. Helix 7 Helix 8 Helix 9 Helix 10 Helix 11
HuGLUT1 VFYYSTSIFEKAGVQQP--VYATIGSGIVNTAFTVVSLFVVERAGRRTLHLIGLAGMAGCAILMTIALALLEQLPWMSYLSIVAIFGFVAFFEVGPGPIPWFIVAELFSQGPRPAAIAVA 407 HuGLUT4 VFYYSTSIFETAGVGQP--AYATIGAGVVNTVFTLVSVLLVERAGRRTLHLLGLAGMCGCAILMTVALLLLERVPAMSYVSIVAIFGFVAFFEIGPGPIPWFIVAELFSQGPRPAAMAVA 423 HuGLUT3 VFYYSTGIFKDAGVQEP--IYATIGAGVVNTIFTVVSLFLVERAGRRTLHMIGLGGMAFCSTLMTVSLLLKDNYNGMSFVCIGAILVFVAFFEIGPGPIPWFIVAELFSQGPRPAAMAVA 405 HuGLUT2 IFYYSTSIFQTAGISKP--VYATIGVGAVNMVFTAVSVFLVEKAGRRSLFLIGMSGMFVCAIFMSVGLVLLNKFSWMSYVSMIAIFLFVSFFEIGPGPIPWFMVAEFFSQGPRPAALAIA 439 ACYPI005975 VFYYSTSLFITAGLAENVAKFVTIGIGVIMVNMTLVTMPLMDKTGRRTLHLYGLGGMFIFSIFITISLLITEMIDWMSYLAVVSILGFVVFFAVGPGSIPWMITAELFSQGPRPAAMSIA 445 DmGLUT1 VFYYSTSLFMSSGLTEESAKFATIGIGAIMVVMTLVSIPLMDRTGRRTLHLYGLGGMFIFSIFITISFLIKEMIDWMSYLSVVATLGFVVFFAVGPGSIPWMITAELFSQGPRPSAMAIA 410 ACYPI001980 VMFFSTKIFNMAGMSNDGAKYATLGMGSLNVIMTLISLFLVELTGRKTLLMIGFSSMFVVTVMLTIALMFVNVSSIVSGLAVVLVMAFVIAFAVGPGSIPWFLVSELFNSSARPLATSIA 425 AB550003 VIFFSTSIFQLASLG-DSAQLATLAMGAMNVLMTVISLVLVERVGRKVLLLVGFSGMFVITCLLAVALAYVKSNKWLPYVCILLVIAFVVMFAVGPGSIPWFLVSELFNQSALPLATSLA 438 AB549996 VIYYSTQIFQKAGMSQQEAQLATMIMGTVNIIMTVISVFLVEIAGRKTLLLIGFGLMFIVTALLAVLLEFIQYD-FASYMCVALVVLFIVCFATGPGSIPWFLVAELFGQDARPLAASIS 405 ::::** :* :.: .*: * : :* ::: ::: .**: * : *:. * : :::: : . . :.: . *: * ***.***::.:*:*.... * * ::: Helix 11 Helix 12
HuGLUT1 GFSNWTSNFIVGMCFQYVEQLCGPYVFIIFTVLLVLFFIFTYFKVPETKGRTFDEIASGFRQG--GASQSDKTPEELFHPLGADSQV------492 HuGLUT4 GFSNWTSNFIIGMGFQYVAEAMGPYVFLLFAVLLLGFFIFTFLRVPETRGRTFDQISAAFHRTP-SLLEQEVKPSTELEYLGPDEND------509 HuGLUT3 GCSNWTSNFLVGLLFPSAAHYLGAYVFIIFTGFLITFLAFTFFKVPETRGRTFEDITRAFEGQAHGADRSGKDGVMEMNSIEPAKETTTNV------496 HuGLUT2 AFSNWTCNFIVALCFQYIADFCGPYVFFLFAGVLLAFTLFTFFKVPETKGKSFEEIAAEFQKKSGSAHRP--KAAVEMKFLGATETV------524 ACYPI005975 VLINWVANFAVGIGFQPLKTALDNYTFLPFSVLLAIFWIFTYKKVPETKNKTFEEILALFRQNGRGSVLESSRLYGTSTTLSDGPGGVCCMRQHWQFPHDDVSERNSPVESHAQ 559 DmGLUT1 VLVNWMANFVVGIGFPSMKTALENYTFLPFSVFLAIFWIFTYKKVPETKNKTFEEILALFRHN------NGSRA------478 ACYPI001980 VGVNWTANFVVGLGFLPLQEMLQSNVFLIFVVLLALFVLYVYKKVPETKNKTLEEIQMGFRQESYK------491 AB550003 VGTNWTANFFVGLGFLPLQQLLGGHVFFIFAILQALFIVFIYKKVPETKNKTLEEISTMFKQISYT------504 AB549996 IGCNWTANFLVGLFFLPLQELIGPKVFIIFAVLQLIFTIFIFFKVPETKNKSLDEVLKYFK------466 14 ** .** :.: * .*: * . * : : :****:.:::::: *.
15 Figure S2. Sequence alignment and transmembrane predictions for GLUT
16 class I transporters. Human GLUT class I transporters (GLUTs 1 - 4) and related
17 transporters from: Acyrthosiphon pisum, Drosophila melanogaster and Nilaparvata
18 lugens are shown. Sequences were aligned using the ClustalX program, and ordered
19 according to their similarity. Membrane spanning regions (Helix 1 – 12) for each
20 transporter were predicted using the TMHMM program
21 (http://www.cbs.dtu.dk/services/TMHMM/) and shaded grey. Boxes highlight
22 conserved Gln177 and the 295MLC motif of ApST4 (ACYPI001980). Asterisks, identical
23 residues in all sequences; colon, conservative amino acid substitution; dot, semi-
24 conservative amino acid substitution.
2 Helix 7
GLUT1 AYRQPILIAVVLQLSQQLSGINAVFYYST 295 GLUT2 SYRQPILVALMLHVAQQFSGINGIFYYST 327 GLUT3 SYRQPIIISIVLQLSQQLSGINAVFYYST 293 GLUT4 THRQPLIIAVVLQLSQQLSGINAVFYYST 311 GLUT14 SYRQPIIISIVLQLSQQLSGINAVFYYST 317 ACYPI005975 TLRQPLIISVVMQLSQQLSGINAVFYYST 331 TC002929 TLRAPLIIGVVMQLSQQLSGINAVFYYST 494 PHUM563670 TLRAPLIIGVVMQLSQQLSGINAVFYYST 509 CG1086_GLUT1 TLRPPLIIGIVMQLSQQFSGINAVFYYST 296 GB13516 TLRAPLIIGVVMQLSQQLSGINAVFYYST 383 TC013487 VMSNVLICCCIINVANQLSGKNALTYFSV 289 TC014313 SLKIPLFIAMLVMVAQQFSGINIVIFYST 291 AB549996 SLRKPLIIAIVIMIAQQLSGINAVIYYST 292 TC013486 ALRIPLIICLCVMIAQQLSGINAVIFFST 319 ACYPI001980 MLKTPLGISVMIMLCQQLSGINAVMFFST 311 PHUM247290 ALRIPLIIAIVVMIGQQLSGINAVMFFST 289 AB550003 ALRIPLFISLVVMIAQQLSGINAVIFFST 325 LOC409424 TLRIPLIIALMVMFAQQLSGINAVMFFST 384 ACYPI009181 SSRKAIIITCIIMLGQQLSGINAVFYYST 340 TC012857 SLLLPLLLVCSLQAGQQFSGINAVFYYSV 306 GB18232 NLKLPIFLVCIIQFGQQMSGINVVFYYSN 324 CG17976_sut3 KLKLPLFIVCSFHFVQQMSGISAIWFYSI 293 CG17975_sut2 ELWLPLLLVCSFQATQQLSGINAIFFYSL 306 CG8714_sut1 RLTLPLIIVCCFHGGQQLSGINAIFYYSV 324 CG7882 QLRLPLIIVCAFLGGQQLSGINAIFYYSV 326 GLUT5 SLRWQLLSIIVLMGGQQLSGVNAIYYYAD 501 GLUT7 SLRWQLLSIIVLMAGQQLSGINAINYYAD 307 GLUT9 YVRWQVVTVIVTMACYQLCGLNAIWFYTN 340 GLUT11 ALRRQVTSLVVLGSAMELCGNDSVYAYAS 304 25 : ::.* . : ::
26 Figure S3. Sequence alignment of TM7 of human GLUT class I and II
27 transporters and insect orthologs. TM7 from human GLUT class I transporters
28 (GLUTs 1-4 and 14); GLUT class II transporters (GLUTs 5, 7, 9 and 11) and all insect
29 orthologs are shown. Sequences from TM7 of all transporters were aligned using
30 the ClustalX program. Boxed regions indicate location of the QLS substrate binding
31 motifs. Functionally characterized transporters are shaded grey and substrate
32 specificities are shown. Transported sugars are abbreviated: frc, fructose; gal,
33 galactose; glc, glucose; man, mannose; myo-ins, myo-inositol. Uncharacterized
34 insect transporters, predicted to be specific for glucose (based on the presence of a
35 QLS motif) are shown in red text. Asterisks indicate identical residues in all
36 sequences; colon indicates conservative amino acid substitution; dot marks semi-
37 conservative amino acid substitution.
3 38 Table S1. Summary of annotated A. pisum sugar porter family transporters.
Gene Nucleotide Protein TIGR00879 score 4 ST ID1 ACYPI ID Scaffold ID2 # TM Helicies3 Identification accession accession (e-value) ApST3 ACYPI004204 LOC100163094 1342 XM_001950662 XP_001950697 12 5.6E-81 ApST1 ACYPI001780 LOC100160486 2 XM_001942918 XP_001942953 12 1.9E-89 ApST4 ACYPI001980 LOC100160702 735 XM_001950928 XP_001950963 12 3.0E-105 ApST7 ACYPI006139 LOC100165178 384 XM_001950568 XP_001950603 12 6.9E-65 ApST7 ACYPI006139 LOC100165178 384 XM_003245156 XP_003245204 12 x ApST26 ACYPI005083 LOC100164041 1482 XM_001949779 XP_001949814 12 5.8E-92 ApST26 ACYPI005083 LOC100164041 1482 XM_003248105 XP_003248153 12 x ApST5 ACYPI003008 LOC100161811 231 XM_001949885 XP_001949920 10 2.8E-62 ApST6 ACYPI009181 LOC100168486 349 XM_001951653 XP_001951688 12 6.6E-80 ApST27 ACYPI009304 LOC100168622 287 XM_001948294 XP_001948329 12 2.2E-68 ApST9 ACYPI001077 LOC100159728 384 XM_001946395 XP_001946430 12 2.2E-77 ApST9 ACYPI001077 LOC100159728 384 XM_003245157 XP_003245205 13 x ApST21 ACYPI006232 LOC100165275 1171 XM_001952605 XP_001952640 10 5.7E-66 ApST16 ACYPI006113 LOC100165150 1 XM_001943769 XP_001943804 12 1.8E-84 ApST28 ACYPI004302 LOC100163202 1171 XM_001952608 XP_001952643 10 8.3E-64 ApST11 ACYPI000812 LOC100159441 1211 XM_001950955 XP_001950990 12 1.1E-90 ApST11 ACYPI000812 LOC100159441 1211 XM_003247820 XP_003247868 12 x ApST43 ACYPI005975 LOC100165001 137 XM_001947713 XP_001947748 12 8.3E-112 ApST43 ACYPI005975 LOC100165001 137 XM_003242843 XP_003242891 12 x ApST43 ACYPI005975 LOC100165001 137 XM_003242844 XP_003242892 12 x ApST43 ACYPI005975 LOC100165001 137 XM_003242845 XP_003242893 12 x ApST43 ACYPI005975 LOC100165001 137 XM_003242846 XP_003242894 12 x ApST17 ACYPI006604 LOC100165672 2 XM_001949996 XP_001950031 10 1.4E-88 ApST17 ACYPI006604 LOC100165672 2 XM_003240048 XP_003240096 10 x ApST22 ACYPI007333 LOC100166465 384 XM_001946496 XP_001946531 10 2.6E-62 ApST23 ACYPI009298 LOC100168616 384 XM_001946444 XP_001946479 12 5.9E-73 ApST24 ACYPI004901 LOC100163845 384 XM_001946310 XP_001946345 10 4.6E-78 ApST25 LOC100165882 384 XM_001946266 XP_001946301 12 1.6E-78 39 ACYPI006799
4 40 Notes: 1A. pisum sugar transporter (ApST) identification number as described in [1]; 2A. pisum genomic scaffold identification
41 number from assembly Acyr_2.0 (NCBI accession number: ABLF00000000.2); 3Transmembrane (TM) helices predicted by
42 TMHMM (available at, http://www.cbs.dtu.dk/services/TMHMM/); 4TIGRFAMS major facilitator superfamily (MFS)
43 transporter, sugar porter (SP) family signature (TIGR00879) e-value scores [2]. Alternative splice forms of ApST genes are
44 shown in light grey.
5 45 Table S2. de novo identification of sugar porter (SP) family transporters
46 [transporter classification number (T.C #) 2.A.1.1] in insect genomes
# of MFS transporter, sugar porter Organism Taxonomy ID Assembly Genebuild family (TIGR00879) transporters Acyrthosiphon pisum 7029 Acyr_2.0 2013-07-AphidBase 19 Anopheles gambiae 7165 AgamP3 2012-10-VectorBase 16 Apis mellifera 7460 Amel4.0 2013-07-BeeBase 8 Drosophila melanogaster 7227 BDGP 5 2012-04-FlyBase 14 Nasonia vitripennis 7425 Nvit_2.1 2010-12-NasoniaBase 11 Pediculus humanus 121224 PhumU1 2009-07-VectorBase 10 Tribolium castaneum 7070 Tcas3 2012-09-24 26 47 Note
48 s: Sugar porter (SP) family transporters (TIGRFAMS motif: TIGR00879 [2]) were
49 identified in insect reference protein datasets (available from
50 http://ensemblgenomes.org/) using HMMER 3.0 motif searches [3]. All identified
51 transporters have a sequence score >237.80 (trusted cutoff).
6 52 Table S3. Summary of annotated A. pisum solute:sodium symporter (SSS) family transporters.
Gene Nucleotide Protein TM PF00474 3 ACYPI ID TCDB top Blast hit Identification accession accession Helicies1 score2 Species Gene symbol Gene name e-value ACYPI009891 LOC100169257 XM_003244407 XP_003244455 13 4.5e-46 H. sapiens SLC5A8 sodium/monocarboxylate cotransporter e-112 ACYPI009891 LOC100169257 XM_001949169 XP_001949204 13 4.5e-46 H. sapiens SLC5A8 sodium/monocarboxylate cotransporter e-112 ACYPI007408 LOC100166547 XM_001951323 XP_001951358 13 1.2e-45 H. sapiens SLC5A8 sodium/monocarboxylate cotransporter e-101 ACYPI000102 LOC100158662 XM_001944878 XP_001944913 13 6.5e-38 H. sapiens SLC5A12 sodium/monocarboxylate cotransporter e-66 ACYPI002047 LOC100160776 XM_001943548 XP_001943583 13 1e-36 H. sapiens SLC5A12 sodium/monocarboxylate cotransporter e-69 ACYPI003487 LOC100162329 XM_001949016 XP_001949051 13 2.7e-36 H. sapiens SLC5A12 sodium/monocarboxylate cotransporter e-66 ACYPI001053 LOC100159703 XM_003241448 XP_003241496 13 3.8e-35 H. sapiens SLC5A8 sodium/monocarboxylate cotransporter e-90 ACYPI001053 LOC100159703 XM_001943453 XP_001943488 13 3.8e-35 H. sapiens SLC5A8 sodium/monocarboxylate cotransporter e-90 53 ACYPI009916 LOC100169284 XM_001944561 XP_001944596 13 3.9e-33 Rattus norvegicus SLC5A7 sodium/choline cotransporter e-151
54 1Transmembrane (TM) helices predicted by TMHMM (available at, http://www.cbs.dtu.dk/services/TMHMM/); 2Pfam E-value
55 score for sodium:solute symporter signature PF00474; 3 Sequence information of top BLASTP hit, aphid transporter versus
56 transporter classification database (TCDB). Alternative splice forms are shown in light grey.
57
58
59
60
61
62
63
7 64 Table S4. Quantitative PCR primers for Acyrthosiphon pisum sugar
65 transporters and housekeeping gene glyceraldehyde-3-phosphate
66 dehydrogenase (GAPDH).
Amplicon QPCR Gene Identification Primer Primer sequence (5’ - 3’) (bp) efficiency (%) ApST1 fwd CCCGGCACAAATCAAAGGT 68 98 (ACYPI001780, LOC100160486) rev CAATGAAAGCGAAGAACCAGTTG ApST3 fwd CGCTGTTGTTTACGGCGTTT 75 93 (ACYPI004204, LOC100163094) rev CGTGCAATATGTCCTGGATTTC ApST4 fwd GCGGTGTGATATCTGGCAGAA 69 93 (ACYPI001980, LOC100160702) rev CGGCCCATATCAATGTCACTT ApST5 fwd GCTATCGTCGGATGGTTGATG 70 104 (ACYPI003008, LOC100161811) rev CCAGGAATCTTCCCACGTAAAT ApST6 fwd AACAATGGTGTAATGAAACGATCATAC 106 91 (ACYPI009181, LOC100168486) rev CCACCAATCAAGAATACGGAAAC ApST7 fwd CCGCATGTTCGCGTTCA 65 97 (ACYPI006139, LOC100165178) rev GCATGCGCCAGAAAAGGA ApST9 fwd CGGACATATTCGGCAGGAA 69 96 (ACYPI001077, LOC100159728) rev TCATTGCCCAGCTGATGATG ApST11 fwd TGCTCAATGGTAGTTGGATTCG 69 99 (ACYPI000812, LOC100159441) rev GCCCGGTCTGTTCATCGA 1 fwd GAGAAAAAGTTCCAATCATGAAAGC ApST16 80 99 (ACYPI006113, LOC100165150) rev AACCATAACCCCAAGGCCTAA 1 fwd GAGAAAAAGTTCCAATCATGAAAGC ApST17 80 94 (ACYPI006604, LOC100165672) rev AACCATAACCCCAAGGCCTAA 2 fwd CGCTCATACTGCCGGATAATG ApST21 74 98 (ACYPI006232, LOC100165275) rev GACCATAGTGACGCCGAACA 2 fwd CGCTCATACTGCCGGATAATG ApST28 74 91 (ACYPI004302, LOC100163202) rev GACCATAGTGACGCCGAACA ApST22 fwd TTGGCGAAGGTGACGATAATT 63 99 (ACYPI007333, LOC100166465) rev CCCAGCGCCGGACAT ApST23 fwd CAGCTTGGGCGTTTACATCA 75 92 (ACYPI009298, LOC100168616) rev AAGAGCTCTCCGACCATGACA ApST27 fwd TCCGCGATCGCATCGT 68 95 (ACYPI009304, LOC100168622) rev ATCGCGTACGTGGCCATAAG ApST43 fwd TCGGCCTGCGGCTATG 63 94 (ACYPI005975, LOC100165001) rev ATGCCGACGGCGAAGTT GAPDH fwd CAATGGAAACAAGATCAAGGTGTT 67 97 67 (ACYPI009769, LOC100169122) rev ACCAGCAGATCCCCATTGG
68 Notes: Due to high sequence similarity between closely related sugar transporters
69 (shaded light grey) it was not possible to design gene specific primers. Primers
70 were designed to co-amplify 1ApST16 and 1ApST17 and co-ampify 2ApST21 and
71 2ApST28.
8 72 Table S5. A. pisum sugar transporter coding sequence primers for
73 Saccharomyces cerevisiae expression constructs.
Amplicon Gene Identification Primer RE site Primer sequence (5’ - 3’) (bp) ApST1 fwd XhoI AACTCGAGATAATGGCTTCAGAAAAAATCGCC 1488 (ACYPI001780, LOC100160486) rev NotI AAGCGGCCGCGTATTTTTCTTTGGTGTCGATGG ApST4 fwd XhoI ATCTCGAGATAATGGCCGAAAAACAAATGGTG 1494 (ACYPI001980, LOC100160702) rev NotI ATGCGGCCGCTTTGTACGATTCCTGTCTGAAACC ApST9 fwd EcoRV ATGATATCATAATGGACCAGGGCGTGTT 1777 (ACYPI001077, LOC100159728) AAGCGGCCGCGACGATCGCGGCGTG 74 rev NotI
75 Notes: Restriction enzyme (RE) sites for forward (fwd) and reverse (rev) primers
76 are underlined in primer sequence and Kozak translation initiation sequences [4]
77 are shown in bold.
9 78 References
79 1. Price DRG, Tibbles K, Shigenobu S, Smertenko A, Russell CW, Douglas AE,
80 Fitches E, Gatehouse AMR, Gatehouse JA: Sugar transporters of the major
81 facilitator superfamily in aphids; from gene prediction to functional
82 characterization. Insect Molecular Biology 2010, 19:97-112.
83 2. Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O:
84 TIGRFAMs: a protein family resource for the functional identification of
85 proteins. Nucleic Acids Research 2001, 29(1):41-43.
86 3. Eddy SR: Profile hidden Markov models. Bioinformatics 1998, 14(9):755-
87 763.
88 4. Kozak M: Structural features in eukaryotic mRNAs that modulate the
89 initiation of translation. Journal of Biological Chemistry 1991,
90 266(30):19867-19870.
91
92
10