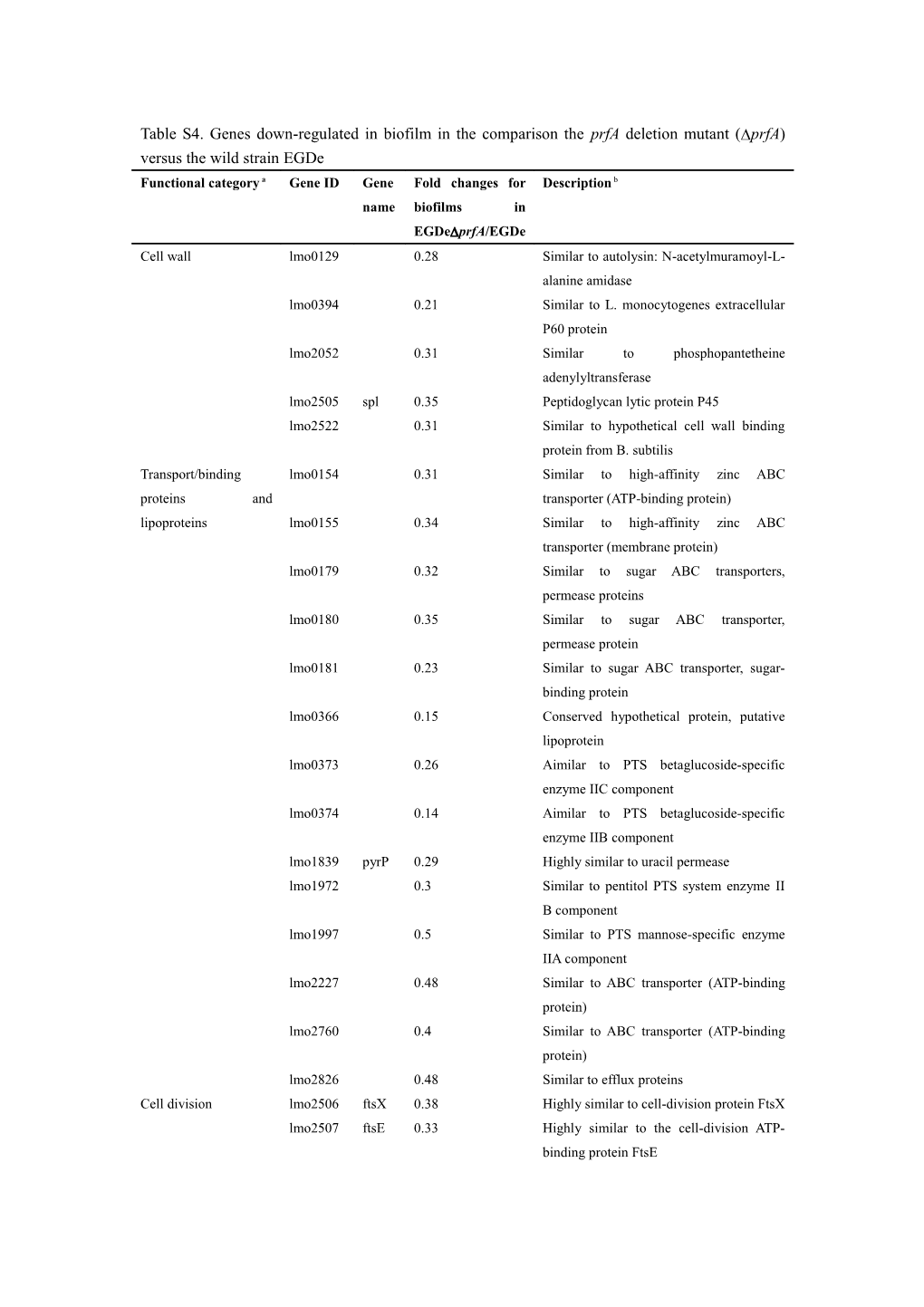
Table S4. Genes down-regulated in biofilm in the comparison the prfA deletion mutant (prfA) versus the wild strain EGDe Functional category a Gene ID Gene Fold changes for Description b name biofilms in EGDeprfA/EGDe Cell wall lmo0129 0.28 Similar to autolysin: N-acetylmuramoyl-L- alanine amidase lmo0394 0.21 Similar to L. monocytogenes extracellular P60 protein lmo2052 0.31 Similar to phosphopantetheine adenylyltransferase lmo2505 spl 0.35 Peptidoglycan lytic protein P45 lmo2522 0.31 Similar to hypothetical cell wall binding protein from B. subtilis Transport/binding lmo0154 0.31 Similar to high-affinity zinc ABC proteins and transporter (ATP-binding protein) lipoproteins lmo0155 0.34 Similar to high-affinity zinc ABC transporter (membrane protein) lmo0179 0.32 Similar to sugar ABC transporters, permease proteins lmo0180 0.35 Similar to sugar ABC transporter, permease protein lmo0181 0.23 Similar to sugar ABC transporter, sugar- binding protein lmo0366 0.15 Conserved hypothetical protein, putative lipoprotein lmo0373 0.26 Aimilar to PTS betaglucoside-specific enzyme IIC component lmo0374 0.14 Aimilar to PTS betaglucoside-specific enzyme IIB component lmo1839 pyrP 0.29 Highly similar to uracil permease lmo1972 0.3 Similar to pentitol PTS system enzyme II B component lmo1997 0.5 Similar to PTS mannose-specific enzyme IIA component lmo2227 0.48 Similar to ABC transporter (ATP-binding protein) lmo2760 0.4 Similar to ABC transporter (ATP-binding protein) lmo2826 0.48 Similar to efflux proteins Cell division lmo2506 ftsX 0.38 Highly similar to cell-division protein FtsX lmo2507 ftsE 0.33 Highly similar to the cell-division ATP- binding protein FtsE lmo2802 gidB 0.44 GidB protein Cell surface proteins lmo0204 actA 0.03 Actin-assembly inducing protein precursor lmo2821 0.10 Similar to internalin, Unknown, putative peptidoglycan bound protein (LPXTG motif) Specific pathways lmo0182 0.14 Similar to alpha-xylosidase and alpha- glucosidase lmo0183 0.2 Similar to alpha-glucosidase lmo0184 0.25 Similar to oligo-1,6-glucosidase lmo0191 0.47 Similar to a putative phospho-beta- glucosidase lmo0372 0.45 Similar to beta-glucosidase lmo0432 0.23 Similar to oxidoreductase lmo1969 0.24 Similar to 2-keto-3-deoxygluconate-6- phosphate aldolase lmo2720 0.29 Similar to acetate-CoA ligase Metabolism of amino lmo0203 mpl 0.02 Zinc metalloproteinase precursor acids and related lmo0458 0.04 Similar to hydantoinase molecules lmo0560 0.1 Similar to NADP-specific glutamate dehydrogenase lmo1733 0.36 Similar to glutamate synthase (small subunit) lmo1734 0.4 Similar to glutamate synthase (large subunit) lmo1983 ilvD 0.22 Similar to dihydroxy-acid dehydratase lmo1984 ilvB 0.22 Similar to acetolactate synthase (acetohydroxy-acid synthase) (large subunit) lmo1986 ilvC 0.24 Similar to ketol-acid reductoisomerase (acetohydroxy-acid isomeroreductase) lmo1987 leuA 0.22 Similar to 2-isopropylmalate synthase lmo1988 leuB 0.21 Similar to 3-isopropylmalate dehydrogenase lmo1990 leuD 0.18 Similar to 3-isopropylmalate dehydratase (small subunit) lmo1991 ilvA 0.14 Similar to threonine dehydratase Metabolism of lmo1837 pyrC 0.46 Highly similar to dihydroorotase nucleotides and nucleic lmo1838 pyrB 0.47 Highly similar to aspartate acids carbamoyltransferase Metabolism of lipids lmo1970 0.28 Similar to putative phosphotriesterase related proteins lmo0201 plcA 0.02 Phosphatidylinositol-specific phospholipase c lmo0205 plcB 0.03 Phospholipase C DNA replication lmo2308 0.15 Similar to single-stranded DNA-binding protein DNA lmo2316 0.10 Similar to site-specific DNA- restriction/modification methyltransferase and repair DNA recombination lmo1277 codV 0.49 Similar to integrase/recombinase Regulation lmo0520 0.49 Similar to transcription regulator lmo0770 0.47 Similar to transcriptional regulator (LacI family) lmo0806 0.37 Similar to transcription regulator lmo0948 0.47 Similar to transcription regulator lmo1280 codY 0.42 Highly similar to B. subtilis CodY protein lmo1840 pyrR 0.26 Highly similar to pyrimidine operon regulatory protein lmo1974 0.37 Similar to transcription regulators, (GntR family) lmo2324 0.15 Similar to anti-repressor [Bacteriophage A118] lmo2328 0.14 Similar to transcription regulator lmo2842 0.47 Similar to transcriptional regulator RNA modification lmo2342 0.41 Similar to 16S pseudouridylate synthase Ribosomal proteins lmo0251 rplL 0.47 Ribosomal protein L12 Adaptation to atypical lmo1278 clpQ 0.49 Highly similar to beta-type subunit of the conditions 20S proteasome Detoxification lmo0395 0.32 Weakly similar to blasticidin S- acetyltransferase Phage-related lmo0121 0.32 Similar to bacteriophage minor tail functions proteins lmo0122 0.39 Similar to phage proteins lmo0123 0.42 Similar to protein gp18 from Bacteriophage A118 lmo0127 0.25 Weakly similar to protein gp20 from Bacteriophage A118 lmo0128 0.28 Similar to a protein from Bacteriophage phi-105 (ORF 45) lmo2293 0.43 Protein gp10 [Bacteriophage A118] lmo2295 0.44 Protein gp8 [Bacteriophage A118] lmo2296 0.42 Similar to coat protein [Bacteriophage SPP1] lmo2299 0.38 Putative portal protein [Bacteriophage A118] lmo2303 0.19 Protein gp66 [Bacteriophage A118] lmo2304 0.19 Bacteriophage A118 gp65 protein lmo2306 0.16 Similar to phage protein lmo2313 0.14 Similar to a bacteriophage protein lmo2315 0.12 Similar to protein gp51 [Bacteriophage A118] lmo2317 0.11 Similar to protein gp49 [Bacteriophage A118] lmo2319 0.18 Similar to bacteriophage proteins lmo2322 0.12 Gp44 [Bacteriophage A118] lmo2323 0.12 Gp43 [Bacteriophage A118] lmo2326 0.13 Similar to protein gp41 [Bacteriophage A118] Miscellaneous lmo0117 lmaB 0.49 Antigen B lmo0202 hly 0.01 Listeriolysin O precursor Similar to unknown lmo0119 0.39 Lmo0119 proteins from other lmo0120 0.37 Lmo0120 organisms lmo0190 0.39 Similar to B. subtilis YabH protein lmo0341 0.49 Lmo0341 lmo0370 0.36 Conserved hypothetical protein lmo0397 0.5 Similar to unknown proteins lmo0457 0.05 Similar to unknown proteins lmo0636 0.37 Similar to unknown proteins lmo0845 0.34 Similar to B. subtilis YxjH and YxjG proteins lmo2083 0.31 Lmo2083 lmo2084 0.48 Lmo2084 lmo2226 0.41 Similar to unknown proteins lmo2318 0.12 Lmo2318 lmo2410 0.48 Lmo2410 lmo2670 0.47 Conserved hypothetical protein lmo2759 0.47 Similar to unknown protein No similarity lmo0104 0.32 Lmo0104 lmo0124 0.38 Lmo0124 lmo0125 0.31 Lmo0125 lmo0126 0.3 Lmo0126 lmo1007 0.33 Lmo1007 lmo2269 0.47 Lmo2269 lmo2302 0.2 Lmo2302 lmo2305 0.17 Lmo2305 lmo2309 0.16 Lmo2309 lmo2310 0.25 Lmo2310 lmo2311 0.16 Lmo2311 lmo2312 0.14 Lmo2312 lmo2314 0.12 Lmo2314 lmo2320 0.12 Lmo2320 lmo2327 0.13 Lmo2327 Unknown c lmo1985 ilvH 0.21 Similar to acetolactate synthase (acetohydroxy-acid synthase) (small subunit) lmo2321 0.13 Lmo2321 a Functional category is classed according to http://genolist.pasteur.fr/ListiList/ (Glaser et al., 2001). b Description for gene is obtained from NCBI (nr) Listeria monocytogenes EGD-e database c Unknown means the function of protein encoded by the identified gene in this study was not found within any category in http://genolist.pasteur.fr/ListiList/.
A Functional Category Is Classed According to (Glaser Et Al., 2001)
Total Page:16
File Type:pdf, Size:1020Kb
Recommended publications