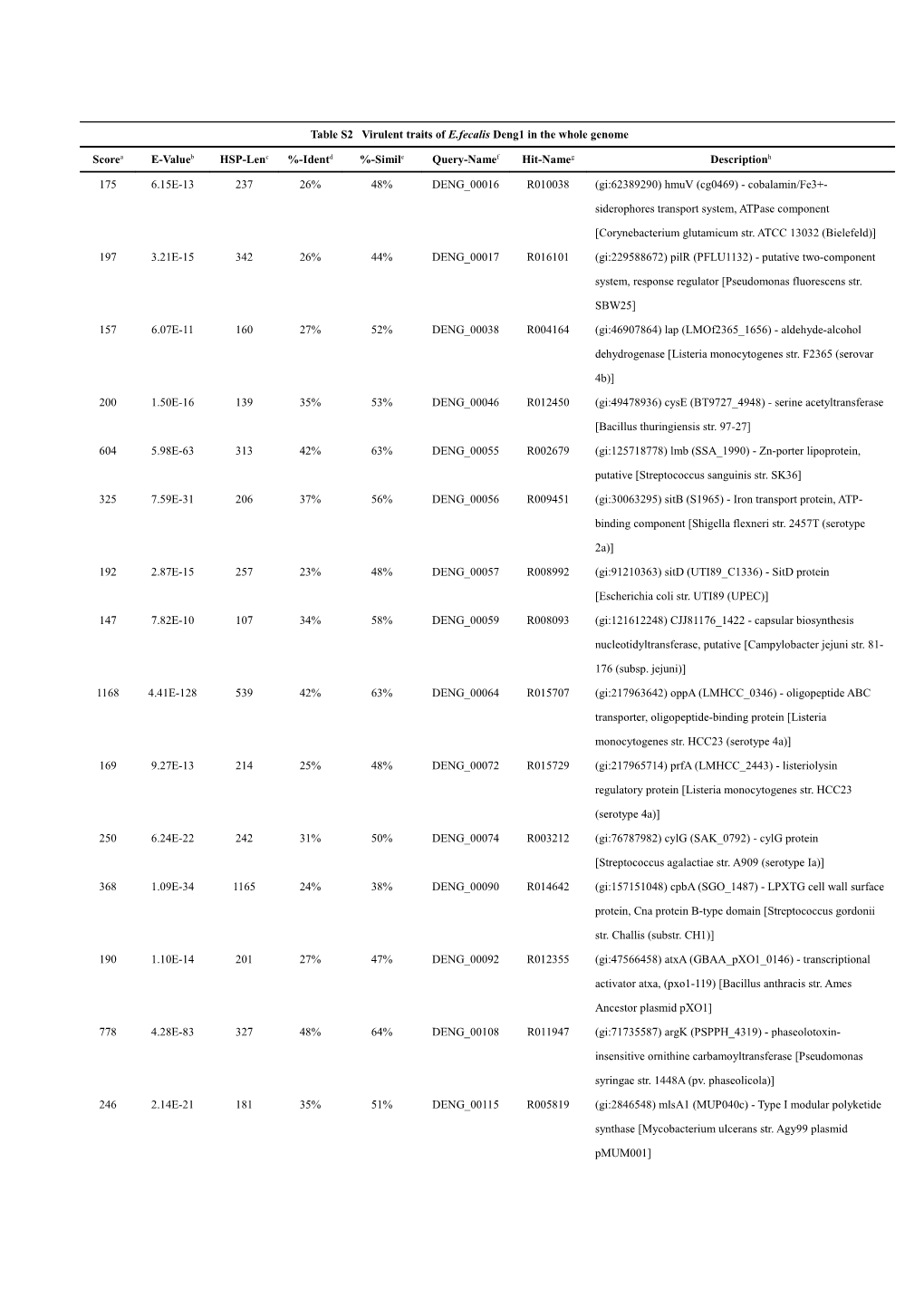
Table S2 Virulent traits of E.fecalis Deng1 in the whole genome
Scorea E-Valueb HSP-Lenc %-Identd %-Simile Query-Namef Hit-Nameg Descriptionh
175 6.15E-13 237 26% 48% DENG_00016 R010038 (gi:62389290) hmuV (cg0469) - cobalamin/Fe3+-
siderophores transport system, ATPase component
[Corynebacterium glutamicum str. ATCC 13032 (Bielefeld)]
197 3.21E-15 342 26% 44% DENG_00017 R016101 (gi:229588672) pilR (PFLU1132) - putative two-component
system, response regulator [Pseudomonas fluorescens str.
SBW25]
157 6.07E-11 160 27% 52% DENG_00038 R004164 (gi:46907864) lap (LMOf2365_1656) - aldehyde-alcohol
dehydrogenase [Listeria monocytogenes str. F2365 (serovar
4b)]
200 1.50E-16 139 35% 53% DENG_00046 R012450 (gi:49478936) cysE (BT9727_4948) - serine acetyltransferase
[Bacillus thuringiensis str. 97-27]
604 5.98E-63 313 42% 63% DENG_00055 R002679 (gi:125718778) lmb (SSA_1990) - Zn-porter lipoprotein,
putative [Streptococcus sanguinis str. SK36]
325 7.59E-31 206 37% 56% DENG_00056 R009451 (gi:30063295) sitB (S1965) - Iron transport protein, ATP-
binding component [Shigella flexneri str. 2457T (serotype
2a)]
192 2.87E-15 257 23% 48% DENG_00057 R008992 (gi:91210363) sitD (UTI89_C1336) - SitD protein
[Escherichia coli str. UTI89 (UPEC)]
147 7.82E-10 107 34% 58% DENG_00059 R008093 (gi:121612248) CJJ81176_1422 - capsular biosynthesis
nucleotidyltransferase, putative [Campylobacter jejuni str. 81-
176 (subsp. jejuni)]
1168 4.41E-128 539 42% 63% DENG_00064 R015707 (gi:217963642) oppA (LMHCC_0346) - oligopeptide ABC
transporter, oligopeptide-binding protein [Listeria
monocytogenes str. HCC23 (serotype 4a)]
169 9.27E-13 214 25% 48% DENG_00072 R015729 (gi:217965714) prfA (LMHCC_2443) - listeriolysin
regulatory protein [Listeria monocytogenes str. HCC23
(serotype 4a)]
250 6.24E-22 242 31% 50% DENG_00074 R003212 (gi:76787982) cylG (SAK_0792) - cylG protein
[Streptococcus agalactiae str. A909 (serotype Ia)]
368 1.09E-34 1165 24% 38% DENG_00090 R014642 (gi:157151048) cpbA (SGO_1487) - LPXTG cell wall surface
protein, Cna protein B-type domain [Streptococcus gordonii
str. Challis (substr. CH1)]
190 1.10E-14 201 27% 47% DENG_00092 R012355 (gi:47566458) atxA (GBAA_pXO1_0146) - transcriptional
activator atxa, (pxo1-119) [Bacillus anthracis str. Ames
Ancestor plasmid pXO1]
778 4.28E-83 327 48% 64% DENG_00108 R011947 (gi:71735587) argK (PSPPH_4319) - phaseolotoxin-
insensitive ornithine carbamoyltransferase [Pseudomonas
syringae str. 1448A (pv. phaseolicola)]
246 2.14E-21 181 35% 51% DENG_00115 R005819 (gi:2846548) mlsA1 (MUP040c) - Type I modular polyketide
synthase [Mycobacterium ulcerans str. Agy99 plasmid
pMUM001] 175 9.35E-13 311 23% 42% DENG_00116 R003015 (gi:28895211) endoS (SPs0299) - hypothetical protein
[Streptococcus pyogenes str. SSI-1 (serotype M3)]
176 1.81E-13 285 29% 42% DENG_00121 R011860 (gi:116049856) phzF2 (PA14_39890) - probable phenazine
biosynthesis protein [Pseudomonas aeruginosa str. UCBPP-
PA14]
452 4.75E-45 531 25% 47% DENG_00125 R008881 (gi:26249408) hlyB (c3573) - Hemolysin B [Escherichia coli
str. CFT073 (UPEC)]
206 7.76E-17 355 25% 45% DENG_00142 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
258 1.54E-22 222 31% 53% DENG_00151 R005294 (gi:120404101) ddrA (Mvan_3122) - daunorubicin resistance
ABC transporter ATPase subunit [Mycobacterium vanbaalenii
str. PYR-1]
190 5.93E-15 346 25% 45% DENG_00164 R010070 (gi:145297067) fagD (cgR_2963) - hypothetical protein
[Corynebacterium glutamicum str. R]
609 1.10E-63 252 46% 67% DENG_00166 R010050 (gi:68536864) fagC (jk1774) - putative iron ABC transport
system, ATP-binding protein [Corynebacterium jeikeium str.
K411]
449 5.31E-45 307 35% 57% DENG_00167 R010065 (gi:62389398) fagA (cg0591) - cobalamin/Fe3+-siderophores
transport system, permease component [Corynebacterium
glutamicum str. ATCC 13032 (Bielefeld)]
440 5.92E-44 335 33% 55% DENG_00168 R010064 (gi:145294652) fagA (cgR_0602) - hypothetical protein
[Corynebacterium glutamicum str. R]
238 2.17E-20 251 30% 47% DENG_00169 R015687 (gi:217964213) lap (LMHCC_0928) - bifunctional
acetaldehyde-CoA/alcohol dehydrogenase [Listeria
monocytogenes str. HCC23 (serotype 4a)]
171 2.48E-12 141 36% 47% DENG_00176 R012525 (gi:13508404) tuf (MPN665) - elongation factor Tu
[Mycoplasma pneumoniae str. M129]
1608 2.62E-179 395 75% 86% DENG_00177 R012532 (gi:42560712) tuf (MSC_0160) - translation elongation factor
Tu [Mycoplasma mycoides str. PG1]
416 4.93E-41 278 34% 51% DENG_00179 R020162 (gi:315444220) pcaA (Mspyr1_26320) - methyltransferase,
cyclopropane fatty acid synthase [Mycobacterium gilvum str.
Spyr1]
331 1.93E-31 237 35% 57% DENG_00214 R008883 (gi:110644661) hlyB (ECP_4554) - alpha-hemolysin
translocation ATP-binding protein HlyB [Escherichia coli str.
536 (UPEC)]
307 1.50E-28 270 32% 55% DENG_00215 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
403 8.90E-40 240 36% 57% DENG_00222 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
228 4.49E-19 224 29% 47% DENG_00223 R000340 (gi:108808062) YPA_2067 - histidine transport system
permease protein HisQ [Yersinia pestis str. Antiqua (biovar
antiqua)] 204 1.02E-16 105 45% 64% DENG_00228 R002520 (gi:21281862) cap8J (MW0133) - capsular polysaccharide
synthesis enzyme Cap8J [Staphylococcus aureus str. MW2]
392 4.18E-38 153 50% 65% DENG_00231 R004173 (gi:16803116) aut (lmo1076) - hypothetical protein [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
135 6.23E-09 102 29% 49% DENG_00260 R010411 (gi:152986357) chpD (PSPA7_0516) - probable
transcriptional regulator [Pseudomonas aeruginosa str. PA7]
117 1.14E-06 246 23% 42% DENG_00266 R003212 (gi:76787982) cylG (SAK_0792) - cylG protein
[Streptococcus agalactiae str. A909 (serotype Ia)]
638 9.58E-67 414 36% 53% DENG_00267 R005651 (gi:145223363) kasB (Mflv_2776) - beta-ketoacyl synthase
[Mycobacterium gilvum str. PYR-GCK]
255 4.84E-23 135 41% 59% DENG_00268 R015527 (gi:163843417) fabZ (BSUIS_A1200) - beta-hydroxyacyl-
(acyl-carrier-protein) dehydratase FabZ [Brucella suis str.
ATCC 23445 chromosome I]
136 7.94E-09 143 30% 50% DENG_00278 R009816 (gi:33152280) kpsF (HD1168) - arabinose-5-phosphate
isomerase [Haemophilus ducreyi str. 35000HP]
318 2.37E-29 606 25% 42% DENG_00283 R014451 (gi:161616855) mgtB (SPAB_04676) - hypothetical protein
[Salmonella enterica str. SPB7 (serovar Paratyphi B)]
355 2.74E-34 225 37% 64% DENG_00359 R004243 (gi:116872809) lisR (lwe1393) - two-component response
regulator [Listeria welshimeri str. SLCC5334 (serovar 6b)]
290 2.47E-26 348 28% 49% DENG_00360 R011397 (gi:146306761) gacS (Pmen_1730) - multi-sensor hybrid
histidine kinase [Pseudomonas mendocina str. ymp]
318 5.18E-30 204 37% 56% DENG_00404 R003702 (gi:15793812) fbpC (NMA0842) - iron-uptake permease ATP-
binding protein [Neisseria meningitidis str. Z2491 (serogroup
A)]
315 1.22E-29 248 33% 52% DENG_00415 R003212 (gi:76787982) cylG (SAK_0792) - cylG protein
[Streptococcus agalactiae str. A909 (serotype Ia)]
143 1.55E-09 120 29% 52% DENG_00420 R016118 (gi:229590420) chpD (PFLU2959) - AraC-family
transcriptional regulator [Pseudomonas fluorescens str.
SBW25]
158 2.50E-11 247 24% 44% DENG_00425 R016118 (gi:229590420) chpD (PFLU2959) - AraC-family
transcriptional regulator [Pseudomonas fluorescens str.
SBW25]
291 1.83E-26 500 22% 48% DENG_00428 R007309 (gi:54293730) iraB (lpl0783) - di/tripeptide transporter
homolog IraB [Legionella pneumophila str. Lens]
346 7.27E-33 499 26% 43% DENG_00432 R016434 (gi:218889443) pchD (PLES_06991) - pyochelin biosynthesis
protein PchD [Pseudomonas aeruginosa str. LESB58]
524 1.78E-53 452 32% 46% DENG_00433 R012194 (gi:70730850) pchA (PFL_3488) - salicylate biosynthesis
isochorismate synthase [Pseudomonas fluorescens str. Pf-5]
118 2.11E-06 176 27% 47% DENG_00438 R011949 (gi:66045851) sypB (Psyr_2615) - Amino acid adenylation
[Pseudomonas syringae str. B728a (pv. syringae)]
140 2.96E-09 155 29% 47% DENG_00447 R008225 (gi:121612579) gmhA-1 (CJJ81176_1166) - phosphoheptose
isomerase [Campylobacter jejuni str. 81-176 (subsp. jejuni)]
430 4.34E-43 201 46% 62% DENG_00451 R006112 (gi:118469767) sodA (MSMEG_6427) - [Mn] superoxide
dismutase [Mycobacterium smegmatis str. MC2 155] 609 1.85E-63 289 43% 63% DENG_00453 R012430 (gi:42784428) lytR (BCE_5383) - membrane-bound
transcriptional regulator LytR [Bacillus cereus str. ATCC
10987]
230 8.46E-20 179 37% 51% DENG_00455 R014454 (gi:161505693) mgtC (SARI_03869) - hypothetical protein
[Salmonella enterica str. RSK2980 (subsp. arizonae serovar
62:z4,z23:--)]
117 5.89E-07 68 38% 62% DENG_00464 R007296 (gi:54298648) feoA (lpp2712) - ferrous iron transporter A
[Legionella pneumophila str. Paris]
985 1.02E-106 727 35% 53% DENG_00465 R007291 (gi:52842863) feoB (lpg2657) - ferrous iron transporter B
[Legionella pneumophila str. Philadelphia 1]
165 2.11E-12 173 29% 49% DENG_00476 R008674 (gi:16132134) fimE (b4313) - tyrosine recombinase/inversion
of on/off regulator of fimA [Escherichia coli str. MG1655
(K12)]
1102 1.27E-120 388 52% 73% DENG_00480 R005070 (gi:27364223) wbfV/wcvB (VV1_0774) - Predicted UDP-
glucose 6-dehydrogenase [Vibrio vulnificus str. CMCP6
chromosome I]
573 2.30E-59 347 37% 61% DENG_00481 R003276 (gi:94993379) hasA (MGAS2096_Spy1882) - Hyaluronan
synthase [Streptococcus pyogenes str. MGAS2096 (serotype
M12)]
191 2.34E-15 220 27% 43% DENG_00484 R009241 (gi:82524546) ipaH2.5 (SDY_P140) - invasion plasmid
antigen, fragment [Shigella dysenteriae str. 197 (serotype 1)
plasmid pSD1_197]
1015 3.45E-110 673 35% 56% DENG_00499 R004541 (gi:147672171) vasG (VC0395_0021) - clpB protein [Vibrio
cholerae str. O395 chromosome I]
1348 2.75E-149 324 74% 88% DENG_00539 R015726 (gi:226224673) bsh (Lm4b_02088) - conjugated bile acid
hydrolase [Listeria monocytogenes str. Clip81459 (serotype
4b)]
154 3.80E-10 80 45% 57% DENG_00547 R003248 (gi:25010541) alp2 (gbs0470) - hypothetical protein
[Streptococcus agalactiae str. NEM316 (serotype III)]
137 1.01E-08 97 26% 54% DENG_00548 R016118 (gi:229590420) chpD (PFLU2959) - AraC-family
transcriptional regulator [Pseudomonas fluorescens str.
SBW25]
110 7.99E-06 136 28% 47% DENG_00557 R009816 (gi:33152280) kpsF (HD1168) - arabinose-5-phosphate
isomerase [Haemophilus ducreyi str. 35000HP]
139 7.10E-10 106 31% 53% DENG_00570 R009241 (gi:82524546) ipaH2.5 (SDY_P140) - invasion plasmid
antigen, fragment [Shigella dysenteriae str. 197 (serotype 1)
plasmid pSD1_197]
321 2.05E-30 189 36% 55% DENG_00617 R004196 (gi:16804235) oppA (lmo2196) - hypothetical protein
[Listeria monocytogenes str. EGD-e (serovar 1/2a)]
445 9.45E-45 230 39% 61% DENG_00622 R008980 (gi:26247467) sitB (c1599) - SitB protein [Escherichia coli
str. CFT073 (UPEC)]
437 1.01E-43 273 35% 62% DENG_00623 R008986 (gi:110641372) sitC (ECP_1190) - iron transport protein,
inner membrane component [Escherichia coli str. 536
(UPEC)] 1284 7.80E-142 300 81% 90% DENG_00624 R003257 (gi:125717136) psaA (SSA_0260) - Manganese/Zinc ABC
transporter substrate-binding lipoprotein precursor, putative
[Streptococcus sanguinis str. SK36]
224 1.07E-18 246 29% 52% DENG_00628 R009584 (gi:16272034) msbA (HI0060) - ABC transporter ATP-
binding protein [Haemophilus influenzae str. Rd KW20
(serotype d)]
695 3.51E-73 558 31% 53% DENG_00630 R012230 (gi:28869794) ybtP (PSPTO_2604) - ABC transporter, ATP-
binding/permease protein [Pseudomonas syringae str. DC3000
(pv. tomato)]
761 7.85E-81 556 35% 55% DENG_00631 R012230 (gi:28869794) ybtP (PSPTO_2604) - ABC transporter, ATP-
binding/permease protein [Pseudomonas syringae str. DC3000
(pv. tomato)]
271 2.07E-24 289 33% 52% DENG_00673 R002016 (gi:49483335) eta (SAR1147) - hypothetical protein
[Staphylococcus aureus str. MRSA252]
406 5.41E-40 324 35% 51% DENG_00674 R016123 (gi:218890650) PLES_19101 - wbpP [Pseudomonas
aeruginosa str. LESB58]
820 5.05E-88 329 48% 69% DENG_00677 R004191 (gi:116872329) lplA1 (lwe0911) - lipoyltransferase and
lipoate-protein ligase family protein [Listeria welshimeri str.
SLCC5334 (serovar 6b)]
171 7.91E-13 122 32% 59% DENG_00682 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
396 6.66E-39 242 38% 58% DENG_00703 R009908 (gi:68248710) hitC (NTHI0180) - iron-utilization ATP-
binding protein hFbpC [Haemophilus influenzae str. 86-
028NP (nontypeable)]
483 1.21E-48 426 32% 48% DENG_00706 R011220 (gi:104784331) algC (PSEEN5434) - phosphomannomutase
[Pseudomonas entomophila str. L48]
333 9.88E-32 220 31% 57% DENG_00724 R005293 (gi:145223975) ddrA (Mflv_3389) - daunorubicin resistance
ABC transporter ATPase subunit [Mycobacterium gilvum str.
PYR-GCK]
633 4.41E-66 410 36% 60% DENG_00737 R008570 (gi:110802269) CPR_0433 - transport protein ysiA
[Clostridium perfringens str. SM101]
169 2.71E-12 173 29% 51% DENG_00738 R012527 (gi:31544430) tuf (MGA_1033) - elongation factor Tu
[Mycoplasma gallisepticum str. R]
1070 1.63E-116 675 39% 56% DENG_00753 R015398 (gi:167033768) clpV1 (PputGB1_2769) - type VI secretion
ATPase, ClpV1 family [Pseudomonas putida str. GB-1]
1426 3.54E-158 428 65% 80% DENG_00764 R002995 (gi:22536290) tig/ropA (SAG0105) - trigger factor
[Streptococcus agalactiae str. 2603V/R (serotype V)]
113 4.90E-06 51 45% 67% DENG_00781 R017583 (gi:194450147) csgD (SeHA_C1253) - DNA-binding
transcriptional regulator CsgD [Salmonella enterica str. SL476
(serovar Heidelberg)]
382 3.55E-37 319 32% 50% DENG_00782 R011947 (gi:71735587) argK (PSPPH_4319) - phaseolotoxin-
insensitive ornithine carbamoyltransferase [Pseudomonas
syringae str. 1448A (pv. phaseolicola)] 184 7.08E-14 189 29% 46% DENG_00810 R017397 (gi:205354641) mgtB (SG3666) - Magnesium transport
ATPase, P-type 2 [Salmonella enterica str. 287/91 (serovar
Gallinarum)]
404 6.55E-40 232 36% 60% DENG_00813 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
195 4.46E-15 212 28% 50% DENG_00814 R000340 (gi:108808062) YPA_2067 - histidine transport system
permease protein HisQ [Yersinia pestis str. Antiqua (biovar
antiqua)]
117 6.42E-06 81 37% 59% DENG_00816 R018132 (gi:300858408) ciuD (cpfrc_00990) - iron ABC transporter
ATP-binding protein [Corynebacterium pseudotuberculosis
str. FRC41]
303 1.11E-27 656 25% 43% DENG_00834 R010420 (gi:15598353) PA3157 - probable acetyltransferase
[Pseudomonas aeruginosa str. PAO1]
152 9.97E-11 255 25% 40% DENG_00839 R018871 (gi:333992762) fbpA (JDM601_4122) - hypothetical protein
[Mycobacterium sp. str. JDM601]
590 5.19E-61 561 30% 55% DENG_00841 R009585 (gi:68248611) msbA (NTHI0072) - lipid A export ATP-
binding protein MsbA [Haemophilus influenzae str. 86-028NP
(nontypeable)]
675 6.26E-71 513 36% 54% DENG_00842 R009587 (gi:148827214) msbA (CGSHiGG_02810) - lipid A export
ATP-binding protein MsbA [Haemophilus influenzae str.
PittGG (nontypeable)]
311 3.50E-29 225 33% 56% DENG_00846 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
432 1.48E-42 177 52% 68% DENG_00854 R004173 (gi:16803116) aut (lmo1076) - hypothetical protein [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
135 1.07E-08 259 22% 43% DENG_00859 R003816 (gi:32266980) peb1 (HH1481) - probable ABC-type amino-
acid transporter periplasmic solute-binding protein
[Helicobacter hepaticus str. ATCC 51449]
379 3.38E-37 204 41% 62% DENG_00860 R014991 (gi:170717476) hitC (HSM_1248) - ABC transporter related
[Haemophilus somnus str. 2336]
232 4.83E-20 211 32% 51% DENG_00861 R000340 (gi:108808062) YPA_2067 - histidine transport system
permease protein HisQ [Yersinia pestis str. Antiqua (biovar
antiqua)]
1019 8.41E-111 536 40% 57% DENG_00862 R004197 (gi:46908430) oppA (LMOf2365_2229) - oligopeptide ABC
transporter, oligopeptide-binding protein [Listeria
monocytogenes str. F2365 (serovar 4b)]
136 5.31E-09 190 28% 54% DENG_00869 R008080 (gi:57238456) CJE1606 - haloacid dehalogenase-like
hydrolase domain/phosphoribulokinase domain protein
[Campylobacter jejuni str. RM1221]
117 1.86E-06 262 23% 39% DENG_00871 R007579 (gi:33591351) bplA (BP0093) - probable oxidoreductase
[Bordetella pertussis str. Tohama I] 435 2.67E-43 263 35% 59% DENG_00912 R014991 (gi:170717476) hitC (HSM_1248) - ABC transporter related
[Haemophilus somnus str. 2336]
120 9.60E-08 61 39% 66% DENG_00916 R012856 (gi:29375148) cylR2 (EF0524) - transcriptional regulator,
Cro/CI family [Enterococcus faecalis str. V583]
752 1.29E-79 920 28% 49% DENG_00918 R014452 (gi:161505694) mgtB (SARI_03870) - hypothetical protein
[Salmonella enterica str. RSK2980 (subsp. arizonae serovar
62:z4,z23:--)]
237 5.04E-20 569 22% 40% DENG_00922 R017399 (gi:194444476) mgtB (SNSL254_A4041) - magnesium-
translocating P-type ATPase [Salmonella enterica str. SL254
(serovar Newport)]
359 2.31E-34 407 27% 48% DENG_00935 R013149 (gi:148269099) icaA (SaurJH9_2690) - glycosyl transferase,
family 2 [Staphylococcus aureus str. JH9]
171 1.03E-12 197 31% 50% DENG_00940 R008131 (gi:15792755) Cj1437c - putative aminotransferase
[Campylobacter jejuni str. NCTC 11168 (subsp. jejuni)]
404 5.49E-40 229 38% 56% DENG_00941 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
277 7.20E-25 220 31% 54% DENG_00944 R000340 (gi:108808062) YPA_2067 - histidine transport system
permease protein HisQ [Yersinia pestis str. Antiqua (biovar
antiqua)]
3277 0 856 70% 83% DENG_00950 R004165 (gi:16800743) lap (lin1675) - hypothetical protein [Listeria
innocua str. Clip11262 (serovar 6a)]
1234 9.99E-136 540 45% 66% DENG_00959 R004198 (gi:16801364) oppA (lin2300) - hypothetical protein [Listeria
innocua str. Clip11262 (serovar 6a)]
334 1.39E-31 165 43% 68% DENG_00964 R012574 (gi:42561491) oppF (MSC_0975) - oligopeptide ABC
transporter, permease component [Mycoplasma mycoides str.
PG1]
412 9.81E-41 177 47% 67% DENG_00965 R012574 (gi:42561491) oppF (MSC_0975) - oligopeptide ABC
transporter, permease component [Mycoplasma mycoides str.
PG1]
651 1.22E-68 224 52% 76% DENG_00981 R004233 (gi:46907975) virR (LMOf2365_1770) - DNA-binding
response regulator [Listeria monocytogenes str. F2365
(serovar 4b)]
519 4.21E-53 330 34% 57% DENG_00982 R004236 (gi:16803781) virS (lmo1741) - hypothetical protein [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
454 1.50E-45 215 41% 64% DENG_00993 R014991 (gi:170717476) hitC (HSM_1248) - ABC transporter related
[Haemophilus somnus str. 2336]
738 3.78E-78 582 32% 56% DENG_00997 R009585 (gi:68248611) msbA (NTHI0072) - lipid A export ATP-
binding protein MsbA [Haemophilus influenzae str. 86-028NP
(nontypeable)]
639 1.04E-66 562 29% 52% DENG_00998 R009588 (gi:33152665) msbA (HD1630) - ABC transporter ATP-
binding protein MsbA [Haemophilus ducreyi str. 35000HP]
1766 0 336 100% 100% DENG_01011 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583] 292 6.68E-27 291 34% 51% DENG_01018 R018987 (gi:333989135) proC (JDM601_0495) - pyrroline-5-
carboxylate reductase ProC [Mycobacterium sp. str. JDM601]
747 1.29E-79 273 54% 74% DENG_01114 R008579 (gi:125973345) Cthe_0827 - hemolysin A [Clostridium
thermocellum str. ATCC 27405]
200 4.54E-16 310 24% 47% DENG_01124 R003288 (gi:24378755) rgpG (SMU.246) - putative glycosyl
transferase N-acetylglucosaminyltransferase), RgpG
[Streptococcus mutans str. UA159]
197 5.56E-16 125 36% 57% DENG_01138 R020325 (gi:387876819) ideR (W7S_17175) - iron-dependent
repressor IdeR [Mycobacterium intracellulare str. MOTT36Y]
245 8.57E-21 223 32% 49% DENG_01143 R004543 (gi:147671674) vasH (VC0395_0020) - sigma-54 dependent
transcriptional regulator [Vibrio cholerae str. O395
chromosome I]
149 3.96E-10 373 22% 38% DENG_01154 R005530 (gi:118473462) eis (MSMEG_3513) - enhanced intracellular
survival protein [Mycobacterium smegmatis str. MC2 155]
123 8.75E-07 118 34% 54% DENG_01165 R009429 (gi:82543034) fepC (SBO_0449) - ATP-binding component of
ferric enterobactin transport [Shigella boydii str. 227 (serotype
4)]
125 4.27E-07 150 25% 46% DENG_01170 R008131 (gi:15792755) Cj1437c - putative aminotransferase
[Campylobacter jejuni str. NCTC 11168 (subsp. jejuni)]
217 6.32E-18 323 25% 44% DENG_01175 R007265 (gi:54294213) lbtB (lpl1278) - hypothetical protein
[Legionella pneumophila str. Lens]
887 5.26E-96 228 80% 90% DENG_01183 R004243 (gi:116872809) lisR (lwe1393) - two-component response
regulator [Listeria welshimeri str. SLCC5334 (serovar 6b)]
874 5.12E-94 492 41% 60% DENG_01184 R004246 (gi:16800483) lisK (lin1415) - two-component sensor
histidine kinase [Listeria innocua str. Clip11262 (serovar 6a)]
398 3.64E-39 208 40% 62% DENG_01189 R014991 (gi:170717476) hitC (HSM_1248) - ABC transporter related
[Haemophilus somnus str. 2336]
1111 1.62E-121 542 40% 64% DENG_01196 R004196 (gi:16804235) oppA (lmo2196) - hypothetical protein
[Listeria monocytogenes str. EGD-e (serovar 1/2a)]
194 1.15E-15 115 38% 57% DENG_01201 R003398 (gi:55821111) eps9 (stu1099) - exopolysaccharide
biosynthesis protein [Streptococcus thermophilus str. LMG
18311 (nonpathogenic)]
1133 2.52E-124 328 62% 77% DENG_01206 R012354 (gi:118480305) galE (BALH_4766) - UDP-glucose 4-
epimerase [Bacillus thuringiensis str. Al Hakam]
210 2.75E-17 357 24% 45% DENG_01208 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
154 3.55E-10 338 27% 44% DENG_01227 R014642 (gi:157151048) cpbA (SGO_1487) - LPXTG cell wall surface
protein, Cna protein B-type domain [Streptococcus gordonii
str. Challis (substr. CH1)]
176 4.40E-13 547 28% 39% DENG_01228 R002724 (gi:25010688) gbs0628 - hypothetical protein [Streptococcus
agalactiae str. NEM316 (serotype III)]
228 5.34E-19 388 28% 40% DENG_01229 R002723 (gi:76787072) SAK_0776 - cell wall surface anchor family
protein [Streptococcus agalactiae str. A909 (serotype Ia)] 489 1.15E-49 287 40% 59% DENG_01230 R015120 (gi:169833610) srtB (SPH_0576) - sortase family protein
[Streptococcus pneumoniae str. Hungary19A-6 (serotype
19A)]
3172 0 674 97% 98% DENG_01235 R012847 (gi:29375675) ace (EF1099) - collagen adhesin protein
[Enterococcus faecalis str. V583]
353 1.34E-33 302 30% 54% DENG_01236 R004437 (gi:15641459) rtxB (VC1448) - RTX toxin transporter [Vibrio
cholerae str. N16961 (O1 biovar eltor) chromosome I]
164 3.79E-12 220 26% 46% DENG_01253 R000340 (gi:108808062) YPA_2067 - histidine transport system
permease protein HisQ [Yersinia pestis str. Antiqua (biovar
antiqua)]
158 2.04E-11 171 26% 49% DENG_01254 R000340 (gi:108808062) YPA_2067 - histidine transport system
permease protein HisQ [Yersinia pestis str. Antiqua (biovar
antiqua)]
655 6.21E-69 271 49% 68% DENG_01255 R008044 (gi:118475479) pebA (CFF8240_1164) - glutamine-binding
protein [Campylobacter fetus str. 82-40 (subsp. fetus)]
351 9.42E-34 236 34% 55% DENG_01257 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
413 1.27E-40 192 48% 62% DENG_01259 R009601 (gi:16272217) orfM (HI0260) - putative deoxyribonucleotide
triphosphate pyrophosphatase [Haemophilus influenzae str.
Rd KW20 (serotype d)]
158 2.00E-11 149 34% 50% DENG_01271 R014623 (gi:161508422) cap5H (USA300HOU_0170) - capsular
polysaccharide biosynthesis protein Cap5H [Staphylococcus
aureus str. USA300_TCH1516]
122 2.30E-07 175 23% 41% DENG_01277 R013545 (gi:153951552) JJD26997_1750 - hypothetical protein
[Campylobacter jejuni str. 269.97 (subsp. doylei)]
200 6.19E-16 235 31% 49% DENG_01316 R008787 (gi:15833822) escN (ECs4568) - EscN [Escherichia coli str.
Sakai (EHEC O157:H7)]
310 2.36E-29 155 45% 60% DENG_01324 R014237 (gi:59711152) luxS (VF0545) - S-ribosylhomocysteinase
[Vibrio fischeri str. ES114 chromosome I]
203 1.25E-16 241 27% 46% DENG_01331 R011932 (gi:71737277) PSPPH_4304 - hydrolase, HAD-superfamily,
subfamily IIA [Pseudomonas syringae str. 1448A (pv.
phaseolicola)]
460 1.78E-46 230 44% 60% DENG_01337 R019258 (gi:333992254) phoP (JDM601_3614) - two-component
system response phosphate regulon transcriptional regulator
PhoP [Mycobacterium sp. str. JDM601]
392 4.72E-38 287 32% 53% DENG_01338 R019259 (gi:183984909) phoR (MMAR_4941) - two-component
system response phosphate sensor kinase, PhoR
[Mycobacterium marinum str. M]
135 8.38E-09 198 28% 43% DENG_01354 R020511 (gi:387873989) phoP (W7S_02915) - DNA-binding response
regulator PhoP [Mycobacterium intracellulare str. MOTT36Y]
634 1.85E-66 277 44% 69% DENG_01356 R012413 (gi:30023297) BC5265 - membrane-bound transcriptional
regulator LytR [Bacillus cereus str. ATCC 14579] 124 1.92E-07 203 25% 44% DENG_01363 R009904 (gi:30995355) hitB (HI0098) - iron(III) ABC transporter
permease protein [Haemophilus influenzae str. Rd KW20
(serotype d)]
167 1.76E-12 202 29% 50% DENG_01364 R003701 (gi:59800669) fbpB (NGO0216) - ABC transporter, permease
protein, iron related [Neisseria gonorrhoeae str. FA 1090]
629 7.94E-66 325 42% 60% DENG_01365 R014991 (gi:170717476) hitC (HSM_1248) - ABC transporter related
[Haemophilus somnus str. 2336]
112 6.33E-06 178 24% 48% DENG_01369 R012363 (gi:118480383) plcR (BALH_4848) - transcription activator
PlcR [Bacillus thuringiensis str. Al Hakam]
143 6.76E-09 382 22% 37% DENG_01384 R015662 (gi:225851651) cgs (BMEA_A0118) - glycosyltransferase 36
[Brucella melitensis str. ATCC 23457 chromosome I]
259 6.37E-23 321 26% 45% DENG_01385 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
128 9.82E-08 192 22% 42% DENG_01390 R007579 (gi:33591351) bplA (BP0093) - probable oxidoreductase
[Bordetella pertussis str. Tohama I]
1461 5.24E-162 565 50% 72% DENG_01395 R002645 (gi:125717748) pavA (SSA_0907) - Fibronectin-binding
protein A, putative [Streptococcus sanguinis str. SK36]
233 4.58E-20 213 26% 56% DENG_01399 R014991 (gi:170717476) hitC (HSM_1248) - ABC transporter related
[Haemophilus somnus str. 2336]
382 1.97E-37 232 39% 59% DENG_01405 R019289 (gi:333989572) mprA (JDM601_0932) - two component
response transcriptional regulatory protein MprA
[Mycobacterium sp. str. JDM601]
318 1.39E-29 530 26% 46% DENG_01406 R011390 (gi:66046927) gacS (Psyr_3698) - Response regulator
receiver:ATP-binding region, ATPase-like:Histidine kinase,
HAMP region:Histidine kinase A, N-terminal:Hpt
[Pseudomonas syringae str. B728a (pv. syringae)]
401 6.11E-39 708 25% 46% DENG_01413 R001477 (gi:62182254) mgtB (SC3684) - Mg2+ transport protein
[Salmonella enterica str. SC-B67 (serovar Choleraesuis)]
569 1.70E-58 482 32% 51% DENG_01415 R014642 (gi:157151048) cpbA (SGO_1487) - LPXTG cell wall surface
protein, Cna protein B-type domain [Streptococcus gordonii
str. Challis (substr. CH1)]
172 2.01E-12 259 33% 48% DENG_01422 R012536 (gi:148377586) tuf (MAG736) - Elongation factor Tu (EF-Tu)
[Mycoplasma agalactiae str. PG2]
205 1.20E-16 156 35% 53% DENG_01442 R015133 (gi:169832565) lytA (SPH_0067) - lytic amidase (N-
acetylmuramoyl-L-alanine amidase) [Streptococcus
pneumoniae str. Hungary19A-6 (serotype 19A)]
111 8.24E-06 88 30% 52% DENG_01453 R003489 (gi:76788234) cpsY (SAK_1263) - transcriptional regulator
CpsY [Streptococcus agalactiae str. A909 (serotype Ia)]
2170 0 902 48% 67% DENG_01456 R014452 (gi:161505694) mgtB (SARI_03870) - hypothetical protein
[Salmonella enterica str. RSK2980 (subsp. arizonae serovar
62:z4,z23:--)]
484 6.03E-49 371 32% 54% DENG_01457 R009963 (gi:68249065) hemN (NTHI0594) - coproporphyrinogen III
oxidase [Haemophilus influenzae str. 86-028NP
(nontypeable)] 148 4.95E-10 64 50% 67% DENG_01463 R011609 (gi:28871892) hopI1 (PSPTO_4776) - type III effector HopI1
[Pseudomonas syringae str. DC3000 (pv. tomato)]
118 1.60E-06 169 25% 49% DENG_01468 R008129 (gi:15792754) Cj1436c - putative aminotransferase
[Campylobacter jejuni str. NCTC 11168 (subsp. jejuni)]
129 5.86E-08 247 27% 47% DENG_01471 R012363 (gi:118480383) plcR (BALH_4848) - transcription activator
PlcR [Bacillus thuringiensis str. Al Hakam]
129 4.38E-08 144 31% 49% DENG_01475 R002873 (gi:71910982) mf3 (M5005_Spy_1169) - streptodornase
[Streptococcus pyogenes str. MGAS5005 (serotype M1)]
322 1.83E-30 216 35% 55% DENG_01476 R009910 (gi:113460924) hitC (HS_0781) - iron(III) ABC transporter,
ATP-binding protein [Haemophilus somnus str. 129PT]
244 2.82E-21 223 25% 48% DENG_01487 R020195 (gi:379762594) ddrA (OCQ_31580) - drrA [Mycobacterium
intracellulare str. MOTT 64]
201 2.24E-16 211 26% 44% DENG_01489 R018795 (gi:183981784) ddrA (MMAR_1771) - daunorubicin-DIM-
transport ATP-binding protein ABC transporter DrrA
[Mycobacterium marinum str. M]
150 7.25E-11 155 27% 43% DENG_01495 R011286 (gi:66043334) algR (Psyr_0063) - Response regulator
receiver:LytTr DNA-binding region [Pseudomonas syringae
str. B728a (pv. syringae)]
317 3.11E-29 201 38% 55% DENG_01501 R009910 (gi:113460924) hitC (HS_0781) - iron(III) ABC transporter,
ATP-binding protein [Haemophilus somnus str. 129PT]
2218 0 876 50% 69% DENG_01511 R017396 (gi:207858995) mgtB (SEN3585) - Magnesium transport
ATPase, P-type 2 [Salmonella enterica str. P125109 (serovar
Enteritidis)]
761 4.03E-81 316 46% 68% DENG_01514 R012544 (gi:47459418) pdhB (MMOB5830) - pyruvate dehydrogenase
E1 component beta subunit [Mycoplasma mobile str. 163K]
670 4.78E-70 727 30% 46% DENG_01552 R013492 (gi:145225066) nuoG (Mflv_4487) - NADH-quinone
oxidoreductase, chain G [Mycobacterium gilvum str. PYR-
GCK]
387 8.92E-38 227 39% 60% DENG_01562 R009910 (gi:113460924) hitC (HS_0781) - iron(III) ABC transporter,
ATP-binding protein [Haemophilus somnus str. 129PT]
135 3.93E-08 144 33% 46% DENG_01563 R014452 (gi:161505694) mgtB (SARI_03870) - hypothetical protein
[Salmonella enterica str. RSK2980 (subsp. arizonae serovar
62:z4,z23:--)]
290 1.46E-26 226 31% 54% DENG_01570 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
147 5.68E-10 361 25% 42% DENG_01572 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
151 3.57E-10 212 26% 41% DENG_01575 R010037 (gi:145294509) hmuV (cgR_0464) - hypothetical protein
[Corynebacterium glutamicum str. R]
176 4.01E-13 388 25% 41% DENG_01580 R009870 (gi:33151641) pgi (HD0418) - glucose-6-phosphate isomerase
[Haemophilus ducreyi str. 35000HP]
326 2.37E-30 311 31% 48% DENG_01662 R004572 (gi:28898442) vscN (VP1668) - type III secretion system ATPase [Vibrio parahaemolyticus str. RIMD 2210633
chromosome I]
368 2.32E-35 350 31% 49% DENG_01663 R010778 (gi:146282926) fliI (PST_2587) - flagellum-specific ATP
synthase FliI [Pseudomonas stutzeri str. A1501]
187 4.92E-15 158 30% 47% DENG_01666 R019231 (gi:169630624) sigH (MAB_3543c) - RNA polymerase
sigma-E factor [Mycobacterium abscessus str. ATCC 19977]
836 1.19E-89 435 42% 59% DENG_01671 R019004 (gi:169628525) lysA (MAB_1434) - diaminopimelate
decarboxylase [Mycobacterium abscessus str. ATCC 19977]
1161 2.90E-127 535 43% 64% DENG_01680 R015707 (gi:217963642) oppA (LMHCC_0346) - oligopeptide ABC
transporter, oligopeptide-binding protein [Listeria
monocytogenes str. HCC23 (serotype 4a)]
445 5.01E-44 694 24% 47% DENG_01685 R014452 (gi:161505694) mgtB (SARI_03870) - hypothetical protein
[Salmonella enterica str. RSK2980 (subsp. arizonae serovar
62:z4,z23:--)]
971 2.16E-105 303 62% 75% DENG_01687 R019211 (gi:253798215) sigA/rpoV (TBMG_01270) - RNA
polymerase sigma factor [Mycobacterium tuberculosis str.
KZN 1435]
996 2.05E-108 337 58% 72% DENG_01693 R015139 (gi:169832817) plr/gapA (SPH_2168) - glyceraldehyde-3-
phosphate dehydrogenase, type I [Streptococcus pneumoniae
str. Hungary19A-6 (serotype 19A)]
617 1.10E-64 250 50% 66% DENG_01701 R015142 (gi:182683690) slrA (SPCG_0720) - peptidyl-prolyl cis-trans
isomerase, cyclophilin-type [Streptococcus pneumoniae str.
CGSP14 (serotype 14)]
169 1.43E-12 168 31% 49% DENG_01704 R008679 (gi:91213958) fimE (UTI89_C5010) - type 1 fimbriae
regulatory protein FimE [Escherichia coli str. UTI89 (UPEC)]
220 2.15E-18 283 28% 49% DENG_01729 R009806 (gi:113461089) kdsA (HS_0946) - 2-dehydro-3-
deoxyphosphooctonate aldolase [Haemophilus somnus str.
129PT]
297 3.06E-27 156 42% 63% DENG_01736 R012430 (gi:42784428) lytR (BCE_5383) - membrane-bound
transcriptional regulator LytR [Bacillus cereus str. ATCC
10987]
187 3.16E-14 275 26% 43% DENG_01742 R010051 (gi:25027240) fagC (CE0684) - putative iron transporter ATP-
binding protein [Corynebacterium efficiens str. YS-314]
408 2.79E-40 154 53% 66% DENG_01751 R004173 (gi:16803116) aut (lmo1076) - hypothetical protein [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
186 1.42E-14 254 29% 44% DENG_01760 R010411 (gi:152986357) chpD (PSPA7_0516) - probable
transcriptional regulator [Pseudomonas aeruginosa str. PA7]
695 3.79E-73 577 30% 54% DENG_01762 R009586 (gi:148825660) msbA (CGSHiEE_03000) - fused lipid
transporter subunits of ABC superfamily: membrane
component/ATP-binding component [Haemophilus influenzae
str. PittEE (nontypeable)]
763 4.35E-81 571 32% 54% DENG_01763 R009585 (gi:68248611) msbA (NTHI0072) - lipid A export ATP-
binding protein MsbA [Haemophilus influenzae str. 86-028NP
(nontypeable)] 128 8.18E-08 302 24% 42% DENG_01768 R012360 (gi:42784526) plcR (BCE_5481) - transcriptional regulator
PlcR, putative [Bacillus cereus str. ATCC 10987]
226 3.82E-19 311 28% 47% DENG_01772 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
1009 1.00E-109 444 45% 66% DENG_01794 R004166 (gi:116873066) lap (lwe1650) - aldehyde-alcohol
dehydrogenase protein [Listeria welshimeri str. SLCC5334
(serovar 6b)]
192 1.38E-15 125 37% 66% DENG_01803 R011285 (gi:28867367) algR (PSPTO_0127) - alginate biosynthesis
regulatory protein AlgR [Pseudomonas syringae str. DC3000
(pv. tomato)]
669 2.29E-70 404 39% 56% DENG_01806 R004163 (gi:16803674) lap (lmo1634) - hypothetical protein [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
392 1.53E-38 225 35% 61% DENG_01811 R016519 (gi:187733815) fepC (SbBS512_E0489) - iron-enterobactin
transporter ATP-binding protein [Shigella boydii str. CDC
3083-94 (serotype 18)]
467 3.78E-47 318 37% 58% DENG_01812 R010028 (gi:38233234) hmuU (DIP0627) - Iron-related transport
system membrane protein [Corynebacterium diphtheriae str.
NCTC 13129 (biotype gravis)]
136 9.31E-09 238 22% 49% DENG_01813 R009008 (gi:15833636) chuT (ECs4382) - putative hemin binding
protein [Escherichia coli str. Sakai (EHEC O157:H7)]
179 1.01E-13 172 29% 48% DENG_01820 R008671 (gi:110644753) fimB (ECP_4646) - type 1 fimbriae
regulatory protein FimB [Escherichia coli str. 536 (UPEC)]
146 6.33E-10 203 27% 49% DENG_01828 R003489 (gi:76788234) cpsY (SAK_1263) - transcriptional regulator
CpsY [Streptococcus agalactiae str. A909 (serotype Ia)]
664 6.70E-70 319 42% 66% DENG_01831 R012544 (gi:47459418) pdhB (MMOB5830) - pyruvate dehydrogenase
E1 component beta subunit [Mycoplasma mobile str. 163K]
243 3.58E-21 255 27% 50% DENG_01840 R003213 (gi:25010702) cylG (gbs0646) - hypothetical protein
[Streptococcus agalactiae str. NEM316 (serotype III)]
107 6.37E-06 125 25% 49% DENG_01841 R008611 (gi:91213834) papX (UTI89_C4886) - putative regulator
PapX protein [Escherichia coli str. UTI89 (UPEC)]
276 5.79E-25 337 33% 48% DENG_01844 R018757 (gi:340627924) ppsC (MCAN_29551) - phenolpthiocerol
synthesis type-I polyketide synthase PPSC [Mycobacterium
canettii str. CIPT 140010059]
235 1.71E-20 191 34% 53% DENG_01846 R014991 (gi:170717476) hitC (HSM_1248) - ABC transporter related
[Haemophilus somnus str. 2336]
217 3.75E-18 215 28% 49% DENG_01852 R018795 (gi:183981784) ddrA (MMAR_1771) - daunorubicin-DIM-
transport ATP-binding protein ABC transporter DrrA
[Mycobacterium marinum str. M]
475 2.76E-48 217 45% 68% DENG_01863 R012281 (gi:15615428) hlyIII (BH2865) - hemolysin III [Bacillus
halodurans str. C-125]
206 6.25E-17 190 30% 54% DENG_01868 R003212 (gi:76787982) cylG (SAK_0792) - cylG protein
[Streptococcus agalactiae str. A909 (serotype Ia)]
177 3.36E-13 198 29% 48% DENG_01877 R010934 (gi:146282910) flhF (PST_2571) - flagellar biosynthesis
protein FlhF [Pseudomonas stutzeri str. A1501] 503 1.70E-51 233 43% 65% DENG_01880 R019253 (gi:169627776) phoP (MAB_0673) - putative DNA-binding
response regulator PhoP [Mycobacterium abscessus str. ATCC
19977]
334 2.19E-31 254 33% 57% DENG_01882 R011387 (gi:152987692) gacS (PSPA7_4582) - sensor/response
regulator hybrid [Pseudomonas aeruginosa str. PA7]
145 1.18E-09 278 24% 45% DENG_01884 R008131 (gi:15792755) Cj1437c - putative aminotransferase
[Campylobacter jejuni str. NCTC 11168 (subsp. jejuni)]
176 3.04E-13 210 27% 48% DENG_01888 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
497 1.55E-50 299 38% 61% DENG_01889 R003489 (gi:76788234) cpsY (SAK_1263) - transcriptional regulator
CpsY [Streptococcus agalactiae str. A909 (serotype Ia)]
118 1.25E-06 170 30% 42% DENG_01898 R011947 (gi:71735587) argK (PSPPH_4319) - phaseolotoxin-
insensitive ornithine carbamoyltransferase [Pseudomonas
syringae str. 1448A (pv. phaseolicola)]
295 9.10E-28 132 52% 71% DENG_01902 R004224 (gi:16803884) lspA (lmo1844) - signal peptidase II [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
729 3.55E-77 576 31% 54% DENG_01910 R009584 (gi:16272034) msbA (HI0060) - ABC transporter ATP-
binding protein [Haemophilus influenzae str. Rd KW20
(serotype d)]
636 2.23E-66 583 30% 52% DENG_01911 R008881 (gi:26249408) hlyB (c3573) - Hemolysin B [Escherichia coli
str. CFT073 (UPEC)]
236 1.63E-20 218 30% 50% DENG_01920 R010050 (gi:68536864) fagC (jk1774) - putative iron ABC transport
system, ATP-binding protein [Corynebacterium jeikeium str.
K411]
349 2.05E-33 328 30% 51% DENG_01921 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
1243 3.80E-137 298 79% 88% DENG_01925 R003327 (gi:71904548) hasC (M28_Spy1886) - UTP-glucose-1-
phosphate uridylyltransferase [Streptococcus pyogenes str.
MGAS6180 (serotype M28)]
668 1.66E-70 265 51% 71% DENG_01927 R004221 (gi:46908654) lgt (LMOf2365_2455) - prolipoprotein
diacylglyceryl transferase [Listeria monocytogenes str. F2365
(serovar 4b)]
302 3.93E-28 247 31% 55% DENG_01934 R009908 (gi:68248710) hitC (NTHI0180) - iron-utilization ATP-
binding protein hFbpC [Haemophilus influenzae str. 86-
028NP (nontypeable)]
283 7.81E-26 241 32% 50% DENG_01935 R010047 (gi:68536376) ciuD (jk1293) - putative iron ABC transport
system, ATP-binding protein [Corynebacterium jeikeium str.
K411]
311 3.22E-29 217 30% 56% DENG_01940 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
1140 1.33E-124 772 37% 54% DENG_01944 R005637 (gi:120404141) secA2 (Mvan_3162) - SecA DEAD domain
protein [Mycobacterium vanbaalenii str. PYR-1] 308 7.75E-29 261 33% 51% DENG_01953 R008022 (gi:83268968) dhbA (BAB2_0012) - Short-chain
dehydrogenase/reductase SDR:Blood group Rhesus C/E and
D polypeptide:Glucose/ribitol dehydrogenase [Brucella
melitensis str. 2308 (biovar Abortus) chromosome II]
127 4.79E-08 88 33% 55% DENG_01962 R007589 (gi:33594847) wbmQ (BPP0128) - putative formyl transferase
[Bordetella parapertussis str. 12822]
119 1.53E-06 185 26% 39% DENG_01964 R007616 (gi:33599134) wbmC (BB0142) - putative glutamine
amidotransferase [Bordetella bronchiseptica str. RB50]
132 2.13E-08 220 24% 45% DENG_01968 R005738 (gi:118471351) purC (MSMEG_5841) -
phosphoribosylaminoimidazole-succinocarboxamide synthase
[Mycobacterium smegmatis str. MC2 155]
980 2.87E-106 483 41% 60% DENG_01974 R015707 (gi:217963642) oppA (LMHCC_0346) - oligopeptide ABC
transporter, oligopeptide-binding protein [Listeria
monocytogenes str. HCC23 (serotype 4a)]
256 6.94E-22 222 35% 52% DENG_01981 R012850 (gi:29375407) EF0818 - polysaccharide lyase, family 8
[Enterococcus faecalis str. V583]
269 3.25E-24 278 28% 49% DENG_01992 R013806 (gi:157415661) C8J_1342 - hypothetical protein
[Campylobacter jejuni str. 81116 (NCTC11828)]
306 1.60E-28 256 32% 50% DENG_01993 R016005 (gi:194100117) lgtC (NGK_2632) - LgtC [Neisseria
gonorrhoeae str. NCCP11945]
1484 4.11E-165 284 99% 99% DENG_01999 R012849 (gi:29376361) sprE (EF1817) - serine proteinase, V8 family
[Enterococcus faecalis str. V583]
2673 0 509 99% 99% DENG_02000 R012848 (gi:29376362) gelE (EF1818) - coccolysin [Enterococcus
faecalis str. V583]
1528 4.02E-170 301 99% 100% DENG_02001 R012853 (gi:29376363) fsrC (EF1820) - histidine kinase, putative
[Enterococcus faecalis str. V583]
212 1.40E-17 215 27% 50% DENG_02013 R009241 (gi:82524546) ipaH2.5 (SDY_P140) - invasion plasmid
antigen, fragment [Shigella dysenteriae str. 197 (serotype 1)
plasmid pSD1_197]
337 1.09E-32 113 56% 74% DENG_02015 R020282 (gi:387873860) panD (W7S_02270) - aspartate alpha-
decarboxylase [Mycobacterium intracellulare str. MOTT36Y]
527 3.96E-54 280 42% 58% DENG_02016 R020274 (gi:315446244) panC (Mspyr1_47470) - pantothenate
synthetase [Mycobacterium gilvum str. Spyr1]
141 3.86E-09 286 22% 45% DENG_02021 R004246 (gi:16800483) lisK (lin1415) - two-component sensor
histidine kinase [Listeria innocua str. Clip11262 (serovar 6a)]
299 7.07E-28 224 34% 57% DENG_02022 R006270 (gi:118473019) mprA (MSMEG_5488) - DNA-binding
response regulator [Mycobacterium smegmatis str. MC2 155]
289 1.20E-26 212 35% 55% DENG_02026 R019136 (gi:169629347) irtB (MAB_2261c) - ABC transporter ATP-
binding protein [Mycobacterium abscessus str. ATCC 19977]
248 1.18E-21 117 46% 62% DENG_02033 R015696 (gi:217965324) iap/cwhA (LMHCC_2049) - invasion
associated secreted endopeptidase [Listeria monocytogenes
str. HCC23 (serotype 4a)]
116 8.59E-06 340 27% 41% DENG_02056 R010009 (gi:38234767) spaG (DIP2227) - probable surface-anchored fimbrial subunit [Corynebacterium diphtheriae str. NCTC
13129 (biotype gravis)]
153 2.32E-10 333 25% 41% DENG_02083 R014972 (gi:170719107) rfaE (HSM_0925) - rfaE bifunctional protein
[Haemophilus somnus str. 2336]
852 3.30E-91 817 30% 53% DENG_02098 R014452 (gi:161505694) mgtB (SARI_03870) - hypothetical protein
[Salmonella enterica str. RSK2980 (subsp. arizonae serovar
62:z4,z23:--)]
188 1.83E-14 261 23% 42% DENG_02104 R009618 (gi:68248881) lpt6 (NTHI0383) - PE-tn-6--
lipooligosaccharide phosphorylethanolamine transferase
[Haemophilus influenzae str. 86-028NP (nontypeable)]
291 3.51E-26 255 35% 50% DENG_02115 R004915 (gi:59712463) flrA (VF1856) - FlrA; sigma-54-dependent
transcriptional activator [Vibrio fischeri str. ES114
chromosome I]
1847 0 433 83% 88% DENG_02118 R003028 (gi:15903080) eno (spr1036) - phosphopyruvate hydratase
[Streptococcus pneumoniae str. R6]
1420 1.30E-157 333 80% 87% DENG_02122 R002805 (gi:15903867) plr/gapA (spr1825) - glyceraldehyde-3-
phosphate dehydrogenase [Streptococcus pneumoniae str. R6]
1598 9.09E-178 717 42% 64% DENG_02132 R006335 (gi:118471771) relA (MSMEG_2965) - GTP
pyrophosphokinase [Mycobacterium smegmatis str. MC2
155]
130 8.10E-09 63 44% 70% DENG_02147 R016497 (gi:187732466) gspG (SbBS512_E3409) - general secretion
pathway protein GspG [Shigella boydii str. CDC 3083-94
(serotype 18)]
140 4.21E-09 356 21% 43% DENG_02148 R006852 (gi:52841753) pilC (lpg1523) - (type IV) pilus assembly
protein PilC [Legionella pneumophila str. Philadelphia 1]
357 2.68E-34 274 31% 55% DENG_02149 R004381 (gi:59710969) mshE (VF0362) - type 4 fimbrial assembly
protein PilB [Vibrio fischeri str. ES114 chromosome I]
306 1.45E-28 208 38% 60% DENG_02153 R003222 (gi:25010705) cylA (gbs0649) - hypothetical protein
[Streptococcus agalactiae str. NEM316 (serotype III)]
231 3.11E-19 242 30% 52% DENG_02155 R014153 (gi:15925989) essC (SA0276) - hypothetical protein
[Staphylococcus aureus str. N315]
476 7.50E-48 522 28% 50% DENG_02159 R009586 (gi:148825660) msbA (CGSHiEE_03000) - fused lipid
transporter subunits of ABC superfamily: membrane
component/ATP-binding component [Haemophilus influenzae
str. PittEE (nontypeable)]
431 1.39E-42 494 27% 48% DENG_02160 R008942 (gi:117624159) ybtQ (APECO1_1055) - putative ABC
transporter protein [Escherichia coli str. O1 (APEC)]
118 1.24E-06 99 29% 55% DENG_02164 R000774 (gi:51593918) virF/lcrF (pYV0076) - lcrF, virF; putative
thermoregulatory protein [Yersinia pseudotuberculosis str.
IP32953 (serotype I) plasmid pYV]
211 3.28E-17 339 26% 43% DENG_02169 R007265 (gi:54294213) lbtB (lpl1278) - hypothetical protein
[Legionella pneumophila str. Lens]
152 1.95E-10 227 26% 45% DENG_02174 R012045 (gi:148546880) pvdN (Pput_1642) - aminotransferase, class V
[Pseudomonas putida str. F1] 491 4.52E-50 251 38% 63% DENG_02176 R009452 (gi:110805357) sitB (SFV_1378) - Iron transport protein,
ATP-binding component [Shigella flexneri str. 8401 (serotype
5b)]
511 3.15E-52 278 39% 63% DENG_02177 R008987 (gi:91210364) sitC (UTI89_C1337) - SitC protein
[Escherichia coli str. UTI89 (UPEC)]
1628 0 308 100% 100% DENG_02178 R012846 (gi:29376585) efaA (EF2076) - endocarditis specific antigen
[Enterococcus faecalis str. V583]
178 2.88E-13 201 27% 49% DENG_02179 R010038 (gi:62389290) hmuV (cg0469) - cobalamin/Fe3+-
siderophores transport system, ATPase component
[Corynebacterium glutamicum str. ATCC 13032 (Bielefeld)]
344 9.17E-33 232 33% 58% DENG_02185 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
151 4.35E-10 168 30% 43% DENG_02192 R007616 (gi:33599134) wbmC (BB0142) - putative glutamine
amidotransferase [Bordetella bronchiseptica str. RB50]
291 2.21E-26 230 30% 52% DENG_02194 R009428 (gi:110804579) fepC (SFV_0536) - ATP-binding component
of ferric enterobactin transport [Shigella flexneri str. 8401
(serotype 5b)]
879 1.09E-94 446 46% 61% DENG_02196 R009851 (gi:148826007) mrsA/glmM (CGSHiEE_04920) - predicted
phosphomannomutase [Haemophilus influenzae str. PittEE
(nontypeable)]
737 3.53E-78 463 37% 55% DENG_02201 R019015 (gi:253798714) glnA1 (TBMG_01760) - glutamine
synthetase glnA1 [Mycobacterium tuberculosis str. KZN
1435]
469 2.37E-47 330 37% 55% DENG_02208 R001468 (gi:29144602) tviC (t4351) - Vi polysaccharide biosynthesis
protein, epimerase [Salmonella enterica str. Ty2 (serovar
Typhi)]
816 2.27E-87 463 38% 60% DENG_02213 R003313 (gi:55823026) epsM (str1076) - exopolysaccharide
biosynthesis protein [Streptococcus thermophilus str.
CNRZ1066 (nonpathogenic)]
179 8.11E-14 245 28% 47% DENG_02215 R012868 (gi:29376985) cpsC (EF2493) - teichoic acid biosynthesis
protein, putative [Enterococcus faecalis str. V583]
294 5.96E-27 369 29% 47% DENG_02217 R012868 (gi:29376985) cpsC (EF2493) - teichoic acid biosynthesis
protein, putative [Enterococcus faecalis str. V583]
442 3.99E-44 217 44% 62% DENG_02218 R003455 (gi:146320426) SSU98_0579 - Glycosyltransferases involved
in cell wall biogenesis [Streptococcus suis str. 98HAH33]
400 2.54E-39 314 34% 54% DENG_02219 R003373 (gi:55823030) epsI (str1080) - exopolysaccharide biosynthesis
protein, sugar transferase [Streptococcus thermophilus str.
CNRZ1066 (nonpathogenic)]
121 9.30E-08 51 47% 65% DENG_02222 R009796 (gi:16273440) licD (HI1540) - lic-1 operon protein
[Haemophilus influenzae str. Rd KW20 (serotype d)]
431 5.06E-43 255 38% 57% DENG_02223 R009690 (gi:16272808) kfiC (HI0868) - hypothetical protein
[Haemophilus influenzae str. Rd KW20 (serotype d)] 403 1.77E-39 376 31% 49% DENG_02224 R005081 (gi:28901258) cpsA (VPA1403) - putative capsular
polysaccharide biosynthesis glycosyltransferase [Vibrio
parahaemolyticus str. RIMD 2210633 chromosome II]
201 9.09E-16 225 29% 49% DENG_02227 R015081 (gi:170025397) YPK_3180 - glycosyl transferase family 2
[Yersinia pseudotuberculosis str. YPIII (serotype O:3)]
200 1.63E-15 122 37% 57% DENG_02228 R003373 (gi:55823030) epsI (str1080) - exopolysaccharide biosynthesis
protein, sugar transferase [Streptococcus thermophilus str.
CNRZ1066 (nonpathogenic)]
537 4.70E-55 309 36% 61% DENG_02229 R003512 (gi:55823379) rgpD (str1469) - polysaccharide ABC
transporter ATP-binding protein [Streptococcus thermophilus
str. CNRZ1066 (nonpathogenic)]
275 6.26E-25 243 28% 52% DENG_02230 R012860 (gi:29376977) cpsK (EF2485) - ABC transporter, permease
protein [Enterococcus faecalis str. V583]
190 8.37E-16 117 38% 63% DENG_02233 R003464 (gi:125718326) SSA_1515 - hypothetical protein
[Streptococcus sanguinis str. SK36]
799 8.49E-86 241 61% 79% DENG_02234 R003526 (gi:116628179) STER_1440 - Glycosyltransferase involved in
cell wall biogenesis [Streptococcus thermophilus str. LMD-9
(nonpathogenic)]
925 2.82E-100 277 62% 77% DENG_02235 R003539 (gi:55821460) rmlD (stu1483) - dTDP-4-keto-L-rhamnose
reductase [Streptococcus thermophilus str. LMG 18311
(nonpathogenic)]
1512 3.03E-168 338 81% 91% DENG_02236 R003509 (gi:55821249) rmlB (stu1242) - dTDP-glucose-4,6-
dehydratase [Streptococcus thermophilus str. LMG 18311
(nonpathogenic)]
463 5.37E-47 181 52% 67% DENG_02237 R005077 (gi:37678487) rmlC (VV0303) - dTDP-6-deoxy-D-xylo-4-
hexulose-3,5-epimerase [Vibrio vulnificus str. YJ016
chromosome I]
1233 5.76E-136 288 80% 88% DENG_02238 R014655 (gi:157151373) rfbA-1 (SGO_1009) - glucose-1-phosphate
thymidylyltransferase [Streptococcus gordonii str. Challis
(substr. CH1)]
298 1.11E-27 219 35% 54% DENG_02239 R003379 (gi:116516251) cps2F (SPD_0321) - glycosyl transferase,
group 2 family protein [Streptococcus pneumoniae str. D39
(serotype 2)]
982 1.03E-106 372 52% 72% DENG_02242 R014665 (gi:157150246) SGO_1723 - RgpG [Streptococcus gordonii
str. Challis (substr. CH1)]
207 5.33E-17 276 25% 43% DENG_02254 R011859 (gi:152986974) phzF2 (PSPA7_3380) - probable phenazine
biosynthesis protein [Pseudomonas aeruginosa str. PA7]
151 3.11E-10 133 29% 48% DENG_02267 R019282 (gi:183984499) mprA (MMAR_4529) - two component
response transcriptional regulatory protein MprA
[Mycobacterium marinum str. M]
210 5.85E-17 200 29% 50% DENG_02268 R011299 (gi:66043335) algZ (Psyr_0064) - Histidine kinase internal
region [Pseudomonas syringae str. B728a (pv. syringae)]
591 3.54E-61 567 30% 50% DENG_02277 R009588 (gi:33152665) msbA (HD1630) - ABC transporter ATP-
binding protein MsbA [Haemophilus ducreyi str. 35000HP] 774 2.09E-82 604 35% 53% DENG_02278 R009585 (gi:68248611) msbA (NTHI0072) - lipid A export ATP-
binding protein MsbA [Haemophilus influenzae str. 86-028NP
(nontypeable)]
160 2.16E-11 351 24% 42% DENG_02291 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
245 2.73E-21 341 25% 42% DENG_02296 R007579 (gi:33591351) bplA (BP0093) - probable oxidoreductase
[Bordetella pertussis str. Tohama I]
151 2.13E-10 193 26% 43% DENG_02297 R007579 (gi:33591351) bplA (BP0093) - probable oxidoreductase
[Bordetella pertussis str. Tohama I]
214 2.19E-17 265 29% 48% DENG_02303 R012527 (gi:31544430) tuf (MGA_1033) - elongation factor Tu
[Mycoplasma gallisepticum str. R]
1307 5.14E-144 719 41% 60% DENG_02306 R016359 (gi:218888835) clpV1 (PLES_00911) - putative ClpA/B-type
chaperone [Pseudomonas aeruginosa str. LESB58]
251 2.36E-22 155 39% 58% DENG_02314 R004173 (gi:16803116) aut (lmo1076) - hypothetical protein [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
123 3.31E-07 240 19% 43% DENG_02317 R007579 (gi:33591351) bplA (BP0093) - probable oxidoreductase
[Bordetella pertussis str. Tohama I]
158 3.52E-11 194 28% 51% DENG_02319 R008129 (gi:15792754) Cj1436c - putative aminotransferase
[Campylobacter jejuni str. NCTC 11168 (subsp. jejuni)]
295 5.92E-27 229 32% 55% DENG_02332 R015367 (gi:167032166) mucP (PputGB1_1153) - membrane-
associated zinc metalloprotease [Pseudomonas putida str. GB-
1]
168 2.62E-12 409 23% 38% DENG_02345 R012047 (gi:70731444) pvdN (PFL_4088) - aminotransferase, class V
[Pseudomonas fluorescens str. Pf-5]
169 1.28E-12 239 25% 51% DENG_02347 R014991 (gi:170717476) hitC (HSM_1248) - ABC transporter related
[Haemophilus somnus str. 2336]
222 8.41E-19 285 31% 48% DENG_02378 R005758 (gi:108797648) proC (Mmcs_0668) - pyrroline-5-carboxylate
reductase [Mycobacterium sp. str. MCS]
815 3.87E-87 583 36% 53% DENG_02379 R009880 (gi:33152559) manB (HD1507) - probable
phosphomannomutase [Haemophilus ducreyi str. 35000HP]
118 1.07E-06 174 26% 43% DENG_02391 R009816 (gi:33152280) kpsF (HD1168) - arabinose-5-phosphate
isomerase [Haemophilus ducreyi str. 35000HP]
255 3.93E-22 273 32% 49% DENG_02419 R012533 (gi:26553484) tuf (MYPE320) - elongation factor Tu
[Mycoplasma penetrans str. HF-2]
142 3.09E-09 197 26% 43% DENG_02420 R008669 (gi:15834525) fimB (ECs5271) - FimB [Escherichia coli str.
Sakai (EHEC O157:H7)]
218 7.13E-18 130 38% 58% DENG_02426 R012532 (gi:42560712) tuf (MSC_0160) - translation elongation factor
Tu [Mycoplasma mycoides str. PG1]
256 1.46E-22 118 48% 66% DENG_02428 R015696 (gi:217965324) iap/cwhA (LMHCC_2049) - invasion
associated secreted endopeptidase [Listeria monocytogenes
str. HCC23 (serotype 4a)]
152 1.83E-10 235 27% 41% DENG_02443 R012592 (gi:42561509) epsG (MSC_0993) - Glycosyltransferase
[Mycoplasma mycoides str. PG1] 153 5.62E-10 594 23% 36% DENG_02445 R010009 (gi:38234767) spaG (DIP2227) - probable surface-anchored
fimbrial subunit [Corynebacterium diphtheriae str. NCTC
13129 (biotype gravis)]
1351 9.91E-150 266 100% 100% DENG_02464 R012869 (gi:29376986) cpsB (EF2494) - phosphatidate
cytidylyltransferase [Enterococcus faecalis str. V583]
1449 4.34E-161 271 100% 100% DENG_02465 R012870 (gi:29376987) cpsA (EF2495) - undecaprenyl diphosphate
synthase [Enterococcus faecalis str. V583]
339 2.98E-32 233 33% 57% DENG_02469 R009910 (gi:113460924) hitC (HS_0781) - iron(III) ABC transporter,
ATP-binding protein [Haemophilus somnus str. 129PT]
143 1.95E-09 305 24% 38% DENG_02505 R012047 (gi:70731444) pvdN (PFL_4088) - aminotransferase, class V
[Pseudomonas fluorescens str. Pf-5]
164 8.18E-12 331 23% 39% DENG_02515 R011947 (gi:71735587) argK (PSPPH_4319) - phaseolotoxin-
insensitive ornithine carbamoyltransferase [Pseudomonas
syringae str. 1448A (pv. phaseolicola)]
837 6.20E-90 320 50% 68% DENG_02528 R012465 (gi:118480308) manA (BALH_4769) - mannose-6-phosphate
isomerase [Bacillus thuringiensis str. Al Hakam]
748 2.41E-79 516 34% 56% DENG_02531 R009585 (gi:68248611) msbA (NTHI0072) - lipid A export ATP-
binding protein MsbA [Haemophilus influenzae str. 86-028NP
(nontypeable)]
627 2.75E-65 559 30% 53% DENG_02532 R009586 (gi:148825660) msbA (CGSHiEE_03000) - fused lipid
transporter subunits of ABC superfamily: membrane
component/ATP-binding component [Haemophilus influenzae
str. PittEE (nontypeable)]
397 9.06E-39 385 29% 51% DENG_02545 R010770 (gi:66046683) fliI (Psyr_3454) - flagellum-specific ATP
synthase [Pseudomonas syringae str. B728a (pv. syringae)]
393 3.45E-38 327 34% 50% DENG_02548 R020062 (gi:332163582) yscN (YE105_P0022) - type III secretion
system ATPase [Yersinia enterocolitica str. 105.5R(r) (subsp.
palearctica) plasmid 105.5R(r)p]
229 3.76E-19 623 24% 41% DENG_02559 R014452 (gi:161505694) mgtB (SARI_03870) - hypothetical protein
[Salmonella enterica str. RSK2980 (subsp. arizonae serovar
62:z4,z23:--)]
367 2.90E-35 177 46% 62% DENG_02563 R004173 (gi:16803116) aut (lmo1076) - hypothetical protein [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
126 1.07E-07 194 27% 45% DENG_02565 R006266 (gi:41407014) mprA (MAP0916) - hypothetical protein
[Mycobacterium avium str. K-10 (subsp. paratuberculosis)]
1939 0 541 71% 84% DENG_02566 R008507 (gi:150015219) groEL (Cbei_0329) - chaperonin GroEL
[Clostridium beijerinckii str. NCIMB 8052]
171 2.19E-12 206 28% 46% DENG_02573 R010037 (gi:145294509) hmuV (cgR_0464) - hypothetical protein
[Corynebacterium glutamicum str. R]
414 7.19E-41 222 37% 63% DENG_02575 R014991 (gi:170717476) hitC (HSM_1248) - ABC transporter related
[Haemophilus somnus str. 2336]
110 9.07E-06 237 23% 46% DENG_02586 R014990 (gi:170717477) hitB (HSM_1249) - binding-protein-
dependent transport systems inner membrane component
[Haemophilus somnus str. 2336] 538 3.00E-55 346 34% 56% DENG_02587 R009910 (gi:113460924) hitC (HS_0781) - iron(III) ABC transporter,
ATP-binding protein [Haemophilus somnus str. 129PT]
114 2.03E-06 133 31% 47% DENG_02607 R007329 (gi:54294502) relA (lpl1571) - GTP pyrophosphokinase
[Legionella pneumophila str. Lens]
775 1.59E-82 576 33% 53% DENG_02619 R009585 (gi:68248611) msbA (NTHI0072) - lipid A export ATP-
binding protein MsbA [Haemophilus influenzae str. 86-028NP
(nontypeable)]
400 6.85E-39 300 36% 59% DENG_02626 R004169 (gi:16802477) inlA (lmo0433) - Internalin A [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
340 2.62E-32 262 34% 53% DENG_02643 R012430 (gi:42784428) lytR (BCE_5383) - membrane-bound
transcriptional regulator LytR [Bacillus cereus str. ATCC
10987]
119 8.17E-07 101 23% 54% DENG_02652 R016118 (gi:229590420) chpD (PFLU2959) - AraC-family
transcriptional regulator [Pseudomonas fluorescens str.
SBW25]
129 1.29E-07 397 20% 43% DENG_02653 R009344 (gi:24111749) gtr (SF0307) - putative glucosyl tranferase II
[Shigella flexneri str. 301 (serotype 2a)]
158 4.94E-11 219 26% 48% DENG_02660 R010053 (gi:62389396) fagC (cg0589) - ABC TRANSPORTER,
NUCLEOTIDE BINDING/ATPASE PROTEIN
[Corynebacterium glutamicum str. ATCC 13032 (Bielefeld)]
1002 6.93E-109 535 39% 60% DENG_02665 R004197 (gi:46908430) oppA (LMOf2365_2229) - oligopeptide ABC
transporter, oligopeptide-binding protein [Listeria
monocytogenes str. F2365 (serovar 4b)]
486 2.41E-49 286 39% 56% DENG_02675 R012864 (gi:29376981) cpsG (EF2489) - MurB family protein
[Enterococcus faecalis str. V583]
185 2.26E-14 326 25% 45% DENG_02677 R010461 (gi:116051142) PA14_23420 - putative zinc-binding
dehydrogenase [Pseudomonas aeruginosa str. UCBPP-PA14]
314 9.54E-30 167 39% 59% DENG_02682 R006083 (gi:41407687) ahpC (MAP1589c) - AhpC [Mycobacterium
avium str. K-10 (subsp. paratuberculosis)]
1089 3.45E-119 335 62% 78% DENG_02684 R004191 (gi:116872329) lplA1 (lwe0911) - lipoyltransferase and
lipoate-protein ligase family protein [Listeria welshimeri str.
SLCC5334 (serovar 6b)]
349 2.76E-33 294 30% 49% DENG_02690 R011174 (gi:146283212) algI (PST_2877) - alginate O-acetylation
protein AlgI [Pseudomonas stutzeri str. A1501]
592 2.44E-61 496 32% 50% DENG_02691 R012088 (gi:104782280) pvdI (PSEEN3229) - pyoverdine sidechain
peptide synthetase [Pseudomonas entomophila str. L48]
290 1.14E-26 247 30% 51% DENG_02694 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
288 1.37E-26 214 34% 53% DENG_02701 R005994 (gi:118469265) irtB (MSMEG_6553) - ABC transporter ATP-
binding protein [Mycobacterium smegmatis str. MC2 155]
234 7.29E-20 204 32% 52% DENG_02713 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945] 1144 1.62E-125 328 63% 78% DENG_02728 R012354 (gi:118480305) galE (BALH_4766) - UDP-glucose 4-
epimerase [Bacillus thuringiensis str. Al Hakam]
234 5.22E-20 220 25% 53% DENG_02751 R007529 (gi:115422956) bvgS (BAV1876) - virulence sensor protein
[Bordetella avium str. 197N]
365 1.87E-35 229 36% 60% DENG_02752 R015621 (gi:163844109) bvrR (BSUIS_A1933) - Transcriptional
regulatory protein chvI [Brucella suis str. ATCC 23445
chromosome I]
326 2.46E-31 144 48% 65% DENG_02776 R015527 (gi:163843417) fabZ (BSUIS_A1200) - beta-hydroxyacyl-
(acyl-carrier-protein) dehydratase FabZ [Brucella suis str.
ATCC 23445 chromosome I]
629 1.11E-65 412 36% 54% DENG_02778 R005650 (gi:118467655) kasB (MSMEG_4328) - 3-oxoacyl-[acyl-
carrier-protein] synthase 2 [Mycobacterium smegmatis str.
MC2 155]
430 5.92E-43 247 38% 62% DENG_02779 R003212 (gi:76787982) cylG (SAK_0792) - cylG protein
[Streptococcus agalactiae str. A909 (serotype Ia)]
346 4.31E-33 298 34% 48% DENG_02780 R005254 (gi:120404102) ppsE (Mvan_3123) - beta-ketoacyl synthase
[Mycobacterium vanbaalenii str. PYR-1]
109 1.96E-06 73 37% 63% DENG_02783 R007841 (gi:23501360) acpXL (BR0459) - acyl carrier protein
[Brucella suis str. 1330 chromosome I]
153 1.38E-10 303 24% 42% DENG_02788 R002485 (gi:73661374) SSP0065 - putative glycosyl transferase
[Staphylococcus saprophyticus str. ATCC 15305]
121 5.64E-07 142 28% 51% DENG_02789 R008198 (gi:118474821) CFF8240_1409 - WabG [Campylobacter fetus
str. 82-40 (subsp. fetus)]
503 1.26E-51 204 49% 70% DENG_02797 R002820 (gi:15900665) slrA (SP_0771) - peptidyl-prolyl cis-trans
isomerase, cyclophilin-type [Streptococcus pneumoniae str.
TIGR4 (serotype 4)]
518 5.94E-53 335 36% 57% DENG_02808 R009910 (gi:113460924) hitC (HS_0781) - iron(III) ABC transporter,
ATP-binding protein [Haemophilus somnus str. 129PT]
654 8.08E-69 309 42% 66% DENG_02809 R009341 (gi:24111748) gtrB (SF0306) - putative bactoprenol glucosyl
transferase [Shigella flexneri str. 301 (serotype 2a)]
372 2.25E-36 211 39% 61% DENG_02812 R020549 (gi:379764654) devR/dosR (OCQ_52190) - two-component
system response regulator [Mycobacterium intracellulare str.
MOTT 64]
189 6.25E-15 204 29% 50% DENG_02813 R006317 (gi:118468427) devS (MSMEG_5241) - GAF family protein
[Mycobacterium smegmatis str. MC2 155]
1115 3.83E-122 365 57% 75% DENG_02819 R003440 (gi:15900286) SP_0357 - UDP-N-acetylglucosamine-2-
epimerase [Streptococcus pneumoniae str. TIGR4 (serotype
4)]
737 5.03E-78 506 34% 58% DENG_02821 R009586 (gi:148825660) msbA (CGSHiEE_03000) - fused lipid
transporter subunits of ABC superfamily: membrane
component/ATP-binding component [Haemophilus influenzae
str. PittEE (nontypeable)]
672 1.62E-70 576 31% 52% DENG_02822 R009587 (gi:148827214) msbA (CGSHiGG_02810) - lipid A export ATP-binding protein MsbA [Haemophilus influenzae str.
PittGG (nontypeable)]
239 7.34E-21 221 30% 54% DENG_02838 R003212 (gi:76787982) cylG (SAK_0792) - cylG protein
[Streptococcus agalactiae str. A909 (serotype Ia)]
169 6.91E-13 115 37% 51% DENG_02839 R003398 (gi:55821111) eps9 (stu1099) - exopolysaccharide
biosynthesis protein [Streptococcus thermophilus str. LMG
18311 (nonpathogenic)]
136 9.84E-09 77 35% 62% DENG_02840 R003489 (gi:76788234) cpsY (SAK_1263) - transcriptional regulator
CpsY [Streptococcus agalactiae str. A909 (serotype Ia)]
117 1.76E-06 302 24% 41% DENG_02844 R009770 (gi:33152291) rfaE (HD1182) - ADP-heptose synthase
[Haemophilus ducreyi str. 35000HP]
236 3.17E-20 345 27% 46% DENG_02845 R012852 (gi:29375537) bopD (EF0954) - sugar-binding transcriptional
regulator, LacI family [Enterococcus faecalis str. V583]
156 4.60E-11 242 29% 44% DENG_02846 R019097 (gi:333990246) mbtJ (JDM601_1606) - acetyl hydrolase MbtJ
[Mycobacterium sp. str. JDM601]
136 1.20E-08 334 21% 40% DENG_02856 R011931 (gi:71733435) PSPPH_4303 - L-arginine:lysine
amidinotransferase, putative [Pseudomonas syringae str.
1448A (pv. phaseolicola)]
269 1.31E-23 239 36% 52% DENG_02867 R004543 (gi:147671674) vasH (VC0395_0020) - sigma-54 dependent
transcriptional regulator [Vibrio cholerae str. O395
chromosome I]
291 6.54E-27 221 33% 52% DENG_02874 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
196 1.35E-15 300 27% 45% DENG_02893 R015727 (gi:217963784) bsh (LMHCC_0491) - choloylglycine
hydrolase (conjugated bile acidhydrolase) (cbah) (bile salt
hydrolase) [Listeria monocytogenes str. HCC23 (serotype 4a)]
743 1.49E-78 894 27% 50% DENG_02904 R017400 (gi:194737976) mgtB (SeSA_A3969) - magnesium-
translocating P-type ATPase [Salmonella enterica str.
CVM19633 (serovar Schwarzengrund)]
150 7.41E-11 156 26% 46% DENG_02913 R019223 (gi:169628453) sigE (MAB_1362) - alternative RNA
polymerase sigma factor [Mycobacterium abscessus str.
ATCC 19977]
7177 0 1372 99% 99% DENG_02916 R012851 (gi:29377483) EF3023 - polysaccharide lyase, family 8
[Enterococcus faecalis str. V583]
1021 4.06E-111 421 54% 69% DENG_02919 R002974 (gi:76788422) htrA/degP (SAK_2135) - serine peptidase HtrA
[Streptococcus agalactiae str. A909 (serotype Ia)]
1143 3.64E-125 539 42% 62% DENG_02926 R004199 (gi:116873629) oppA (lwe2213) - periplasmic oligopeptide-
binding protein [Listeria welshimeri str. SLCC5334 (serovar
6b)]
578 3.58E-60 236 48% 65% DENG_02942 R002826 (gi:24379545) srtA (SMU.1113) - putative sortase
[Streptococcus mutans str. UA159]
169 4.86E-13 149 30% 48% DENG_02945 R005042 (gi:59710771) wzb (VF0164) - Protein tyrosine phosphatase
[Vibrio fischeri str. ES114 chromosome I] 880 1.19E-94 539 35% 58% DENG_02967 R015707 (gi:217963642) oppA (LMHCC_0346) - oligopeptide ABC
transporter, oligopeptide-binding protein [Listeria
monocytogenes str. HCC23 (serotype 4a)]
694 1.86E-73 321 47% 64% DENG_02968 R003260 (gi:15901700) piuA (SP_1872) - iron-compound ABC
transporter, iron-compound-binding protein [Streptococcus
pneumoniae str. TIGR4 (serotype 4)]
712 1.37E-75 251 49% 73% DENG_02970 R004671 (gi:28900515) vctC (VPA0660) - iron(III) ABC transporter,
ATP-binding protein [Vibrio parahaemolyticus str. RIMD
2210633 chromosome II]
495 2.40E-50 315 38% 60% DENG_02971 R004668 (gi:37676279) vctG (VVA0619) - catechol siderophore ABC
transporter, permease protein [Vibrio vulnificus str. YJ016
chromosome II]
714 9.75E-76 317 43% 72% DENG_02972 R004660 (gi:147672165) vctD (VC0395_1000) - vibriobactin and
enterobactin ABC transporter, permease protein [Vibrio
cholerae str. O395 chromosome I]
178 2.19E-13 202 27% 47% DENG_02982 R004031 (gi:32266642) flhF (HH1143) - flagellar biosynthesis protein
[Helicobacter hepaticus str. ATCC 51449]
371 6.47E-36 193 44% 62% DENG_02991 R012574 (gi:42561491) oppF (MSC_0975) - oligopeptide ABC
transporter, permease component [Mycoplasma mycoides str.
PG1]
330 3.56E-31 257 33% 51% DENG_02992 R009910 (gi:113460924) hitC (HS_0781) - iron(III) ABC transporter,
ATP-binding protein [Haemophilus somnus str. 129PT]
154 1.40E-11 68 47% 68% DENG_02993 R007841 (gi:23501360) acpXL (BR0459) - acyl carrier protein
[Brucella suis str. 1330 chromosome I]
553 3.40E-57 242 46% 71% DENG_02998 R004251 (gi:116871464) agrA (lwe0042) - 2-components response
regulator protein, AlgR/VirR/ComE type [Listeria welshimeri
str. SLCC5334 (serovar 6b)]
436 2.54E-43 412 29% 54% DENG_02999 R004253 (gi:46906281) agrC (LMOf2365_0059) - accessory gene
regulator protein C [Listeria monocytogenes str. F2365
(serovar 4b)]
269 3.43E-24 223 30% 50% DENG_03001 R016054 (gi:194097927) fbpC (NGK_0348) - Fe(3+) ions import ATP-
binding protein fbpC [Neisseria gonorrhoeae str.
NCCP11945]
468 8.71E-47 318 36% 51% DENG_03006 R016329 (gi:229593375) ppkA (PFLU6010) - putative serine/threonine
protein kinase [Pseudomonas fluorescens str. SBW25]
589 2.39E-61 243 49% 69% DENG_03007 R004208 (gi:16803861) stp (lmo1821) - hypothetical protein [Listeria
monocytogenes str. EGD-e (serovar 1/2a)]
288 2.26E-26 277 29% 51% DENG_03010 R018197 (gi:118470693) fxbA (MSMEG_0014) - Formyl transferase
[Mycobacterium smegmatis str. MC2 155]
162 3.10E-12 158 29% 47% DENG_03073 R019236 (gi:333991577) sigH (JDM601_2937) - RNA polymerase
sigma-E factor (sigma- 24) SigH (RpoE) [Mycobacterium sp.
str. JDM601]
129 8.25E-08 100 29% 47% DENG_03085 R019092 (gi:169631131) mbtJ (MAB_4052c) - putative lipase/esterase
[Mycobacterium abscessus str. ATCC 19977] 117 7.37E-07 80 38% 51% DENG_03086 R004611 (gi:147674293) vibB (VC0395_A0300) - vibriobactin-
specific isochorismatase [Vibrio cholerae str. O395
chromosome II]
282 8.00E-26 255 31% 51% DENG_03090 R011283 (gi:152989087) algR (PSPA7_6007) - alginate biosynthesis
regulatory protein AlgR [Pseudomonas aeruginosa str. PA7]
246 4.51E-21 209 33% 50% DENG_03091 R011301 (gi:146305319) algZ (Pmen_0278) - histidine kinase internal
region [Pseudomonas mendocina str. ymp]
333 1.08E-31 218 37% 59% DENG_03094 R003702 (gi:15793812) fbpC (NMA0842) - iron-uptake permease ATP-
binding protein [Neisseria meningitidis str. Z2491 (serogroup
A)]
554 7.15E-57 317 40% 62% DENG_03101 R002679 (gi:125718778) lmb (SSA_1990) - Zn-porter lipoprotein,
putative [Streptococcus sanguinis str. SK36]
111 6.00E-06 231 27% 47% DENG_03103 R012860 (gi:29376977) cpsK (EF2485) - ABC transporter, permease
protein [Enterococcus faecalis str. V583]
425 3.55E-42 293 34% 54% DENG_03104 R020192 (gi:406031303) ddrA (MIP_04588) - Daunorubicin resistance
ATP-binding protein drrA [Mycobacterium indicus pranii str.
MTCC 9506]
258 2.45E-22 246 33% 49% DENG_03109 R004543 (gi:147671674) vasH (VC0395_0020) - sigma-54 dependent
transcriptional regulator [Vibrio cholerae str. O395
chromosome I]
330 1.08E-30 256 38% 52% DENG_03110 R002764 (gi:116516664) cbpD (SPD_2028) - choline binding protein D
[Streptococcus pneumoniae str. D39 (serotype 2)]
139 1.93E-08 452 23% 41% DENG_03114 R009617 (gi:16272233) lpt6 (HI0275) - hypothetical protein
[Haemophilus influenzae str. Rd KW20 (serotype d)]
172 1.95E-13 148 28% 51% DENG_03117 R003821 (gi:32265709) napA (HH0210) - starvation-inducible DNA-
binding protein Dps [Helicobacter hepaticus str. ATCC 51449]
121 2.30E-07 158 28% 46% DENG_03121 R015208 (gi:170723699) pilD (PputW619_4538) - Prepilin peptidase
[Pseudomonas putida str. W619]
546 5.30E-56 310 39% 58% DENG_03131 R012430 (gi:42784428) lytR (BCE_5383) - membrane-bound
transcriptional regulator LytR [Bacillus cereus str. ATCC
10987]
264 7.34E-24 188 32% 55% DENG_03154 R009603 (gi:148825461) orfM (CGSHiEE_01765) - putative
deoxyribonucleotide triphosphate pyrophosphatase
[Haemophilus influenzae str. PittEE (nontypeable)]
1216 2.07E-133 840 37% 57% DENG_03171 R015399 (gi:170721686) clpV1 (PputW619_2505) - type VI secretion
ATPase, ClpV1 family [Pseudomonas putida str. W619]
385 9.66E-38 229 37% 60% DENG_03180 R015621 (gi:163844109) bvrR (BSUIS_A1933) - Transcriptional
regulatory protein chvI [Brucella suis str. ATCC 23445
chromosome I]
316 1.71E-29 275 29% 50% DENG_03181 R019259 (gi:183984909) phoR (MMAR_4941) - two-component
system response phosphate sensor kinase, PhoR
[Mycobacterium marinum str. M]
133 1.70E-08 123 34% 46% DENG_03189 R012425 (gi:30023309) BC5277 - Tyrosine-protein kinase (capsular polysaccharide biosynthesis) [Bacillus cereus str. ATCC
14579]
292 6.83E-27 261 33% 51% DENG_03201 R003213 (gi:25010702) cylG (gbs0646) - hypothetical protein
[Streptococcus agalactiae str. NEM316 (serotype III)]
261 2.13E-23 230 30% 51% DENG_03223 R019253 (gi:169627776) phoP (MAB_0673) - putative DNA-binding
response regulator PhoP [Mycobacterium abscessus str. ATCC
19977]
Note: a:Score: The score of the target protein used to blast. b:E-value: stastistic value to blast analysis. c:HSP-Len: the highest protein length which used to be blasted. d:Ident: the identity of the amino acid sequence compared with the homogeneous protein in VFDB database. e:Similarity: the similarityof the amino acid sequence compared with the homogeneous protein in VFDB database. f: Query- name: The numbers similar to the protein in E.fecalis Deng1. g:Hit-Name: the number of the most suitable protein in the VFDB database used to analyze the target protein. For example, R010038 represents the number of the protein used to compared with DENG_0001. h:Description: the specific description of the protein used in VFDB database( http://www.mgc.ac.cn/VFs/main.htm) including the abbreviation of the virulent factors, the number of genebank ID in NCBI data, base locus_tag, and the bacteria strain name which also express this protein.