Supplementary Material Genetic and Biochemical Mechanisms of Pollen Wall Development Jianxin Shi1, Meihua Cui1, Li Yang1, Yu-Jin Kim1,2, Dabing Zhang1,3 1Joint International Research Laboratory of Metabolic & Developmental Sciences, SJTU-University of Adelaide Joint Centre for Agriculture and Health, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, 800 Dongchuan Rd., Shanghai 200240, China 2Department of Oriental Medicinal Biotechnology and Graduate School of Biotechnology, College of Life Science, Kyung Hee University, Youngin, 446-701, South Korea 3School of Agriculture, Food and Wine, University of Adelaide, South Australia 5064, Australia Corresponding author: Zhang, D. ([email protected])
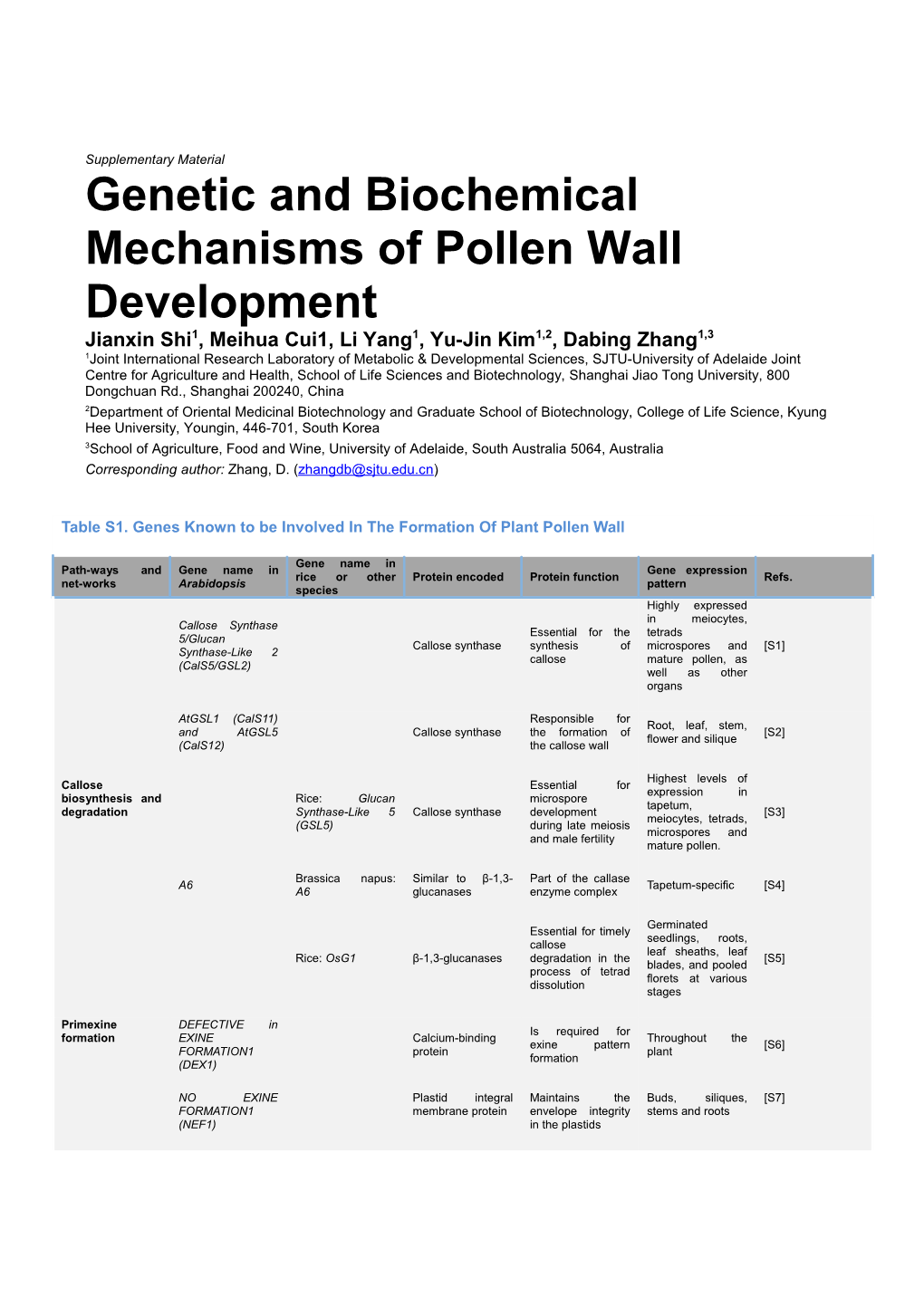
Table S1. Genes Known to be Involved In The Formation Of Plant Pollen Wall
Gene name in Path-ways and Gene name in Gene expression rice or other Protein encoded Protein function Refs. net-works Arabidopsis pattern species Highly expressed in meiocytes, Callose Synthase Essential for the tetrads 5/Glucan Callose synthase synthesis of microspores and [S1] Synthase-Like 2 callose mature pollen, as (CalS5/GSL2) well as other organs
AtGSL1 (CalS11) Responsible for Root, leaf, stem, and AtGSL5 Callose synthase the formation of [S2] flower and silique (CalS12) the callose wall
Highest levels of Callose Essential for expression in biosynthesis and Rice: Glucan microspore tapetum, degradation Synthase-Like 5 Callose synthase development [S3] meiocytes, tetrads, (GSL5) during late meiosis microspores and and male fertility mature pollen.
Brassica napus: Similar to β-1,3- Part of the callase A6 Tapetum-specific [S4] A6 glucanases enzyme complex
Germinated Essential for timely seedlings, roots, callose leaf sheaths, leaf Rice: OsG1 β-1,3-glucanases degradation in the [S5] blades, and pooled process of tetrad florets at various dissolution stages
Primexine DEFECTIVE in Is required for formation EXINE Calcium-binding Throughout the exine pattern [S6] FORMATION1 protein plant formation (DEX1)
NO EXINE Plastid integral Maintains the Buds, siliques, [S7] FORMATION1 membrane protein envelope integrity stems and roots (NEF1) in the plastids Exine pattern Microspores and RUPTURED MtN3/saliva family formation and tapetum POLLEN GRAIN 1 [S8, S9] protein primexine (RPG1) deposition during meiosis
RUPTURED Functions MtN3/saliva family Tapetum and POLLEN GRAIN 1 redundantly with [S9] protein microspores (RPG2) RPG1
Plays a vital role in NO PRIMEXINE primexine AND PLASMA deposition and Microspores and MEMBRANE Membrane protein plasma membrane [S10] the tapetum UNDULATION undulation during (NPU) early pollen wall development.
Required for pollen exine formation and male fertility Microsporocytes, Exine Formation DNA via the regulation tetrads and the [S11] Defect (EFD) methyltransferase of callose wall and tapetum primexine formation
Biosynthesis of Regulates the sporopollenin production of key ATP- AtAPY6 and polysaccharides Mature pollen diphosphohydrolas [S12] AtAPY7 needed for proper grains es assembly of the exine layer
AtACBP4 and AtACBP6: pollen AtACBP4, Affects Acyl-CoA-binding grains. AtACBP5 and sporopollenin and [S13] proteins AtACBP5:microspo AtACBP6 tryphine synthesis res and tapetal cells
Synthesis of arabinan side Rice: UDP- Pistil, anther, UDP- chains of Arabinopyranose palea, lamma, root, Arabinopyranose rhamnogalacturon [S14] Mutase 3 leaf sheath, Mutase an-I is essential for (OsUAM3) internode and leaf pollen wall formation
Tapetum and Rice: DEFECTIVE shortly after MALE STERILITY Fatty acyl carrier Required for pollen POLLEN WALL release of [S15] 2 (MS2) protein reductase exine development (DPW) microspore from tetrad
Catalyzes in-chain Mid-chain fatty acid hydroxylation of Tapetum and CYP703A2 Rice: CYP703A3 [S16, S17] hydroxylase saturated medium- microspore specific chain fatty acids
Catalyzes v- hydroxylation of long-chain fatty Fatty acid ω- CYP704B1 Rice: CYP704B2 acids, implicating Anther-specific [S18, S19] hydroxylase these molecules in sporopollenin synthesis
Acyl-CoA Acyl-CoA synthase Required for Tapetum-specific [S20] Synthase 5 gene sporopollenin (ACOS5) monomer biosynthesis Required for pollen At polyketide Polyketide development and synthases Tapetum-specific [S21] synthases sporopollenin (AtPKSA/LAP5) biosynthesis
Required for pollen At polyketide Polyketide development and synthases Tapetum-specific [S21] synthases sporopollenin (AtPKSB/LAP6) biosynthesis
Tetraketide α- Sporopollenin Tetraketide α- pyrone reductase monomer Tapetum-specific [S22] pyrone reductase 1 1 (TKPR1) biosynthesis
Tetraketide α- Sporopollenin Tetraketide α- pyrone reductase monomer Tapetum-specific [S22] pyrone reductase 2 2 (TKPR2) biosynthesis
Epicuticular wax ECERIFERUM 1 Integral membrane and tryphine Mainly in stem, fruit [S23] (CER1) protein alkane and leaf biosynthesis
Play a role in the synthesis of the components of Buds, open Faceless Pollen 1 Aldehyde tryphine, flowers, siliques, [S24] (FLP1)/CER3 decarboxylase sporopollenin of stems, leaves but exine and the wax not roots of stems and siliques
Tryphine Importance for formation Fatty acid cuticle formation Tapetum and CER2-LIKE2 [S25] elongase and pollen coat microspores function
LACS1 and Long-chain acyl- Production of Anther-specific [S26] LACS4 CoA synthetases tryphine lipids expression
Affects oil bodies formation in the tapetosomes of Phosphoserine tapetalcells which, Microspore and PSP1 [S27] phosphatase in turn, may tapetal cell influence microspore pollen coat formation
Intine Involved in development Fasciclin-like microspore Pollen grains and FLA3 arabinogalactan development and [S28] tubes proteins may affect pollen intine formation
Critical role in Inflorescence, Sugar pyrophos- AtUSP pollen particularly in [S29] phorylase development. pollen grains
Rice: Essential for intine GLYCOSYLTRAN Glycosyl- construction and Mature pollen [S30] SFERASE1 transferases pollen maturation (OsGT1)
Rice: Cell wall COLLAPSED Arabinokinase-like Tapetum and polysaccharides [S31] ABNORMAL protein microspores synthesis POLLEN1 (CAP1)
Brassica Pectatelyase-like Intine and tryphine Microspores and [S32] campestris: 10 formation pistils BcPLL10 Brassica campestris: Pectatelyase-like 9 Intine formation Pollen and pistils [S33] BcPLL9
Modulate the physical nature of Brassica Arabinogalactan the pollen wall and Pollen grains and campestris: [S34] protein the integrity of the pollen tubes BcMF8 pollen tube wall matrix.
Brassica Participate in the BcMF26a: pistil campestris: construction of Polygalacturonase pollen wall by BcMF26b: [S35] BcMF26a and modulating intine tapetum, pollen BcMF26b information grains, and pistils
Affects formation of the pollen intine at Pollen grains at the Brassica oleracea: Polygalacturonase the late stage of mature pollen [S36] BoMF25 pollen stage development
Transfer of sporopollenin lipid ATP binding precursors from ABCG26 cassette tapetal cells to Tapetum [S37, S38] transporter anther locules, facilitating exine formation
ATP binding ABCG9 and Pollen coat cassette Tapetum [S39] ABCG31 deposition transporter
ATP binding ABCG1 and Transport lipid cassette Tapetum [S40] ABCG16 precursors transporter
ATP binding Rice: Sporopollenin Preferentially in the cassette [S41, S42] OsABCG15/PDA1 transfer tapetum transporter Transporters type III LTPs Transport Lipid transfer (LTP6, LTP9 and sporopollenin Tapetum [S43] proteins LTP14) precursors
Plays a crucial role Mainly in tapetum Lipid transfer in the development Rice: OsC6 and weakly in [S44] protein of lipidic orbicules microspore and pollen exine
Magnesium Magnesium transporter and Notably in pollen AtMGT4 [S45] transporter essential for pollen grains development
Biosynthesis REPRESSOR OF sporopollenin or Strongly expressed CYTOKININ Nucleotide sugar polysaccharide- in the shoot apical [S46] DEFICIENCY 1 transporter containing meristem and (ROCK1) primexine young flowers formation
Transcription ABORTED Rice: TAPETUM bHLH transcription A master regulator Tapetum [S47, S48, S49] factors MICROSPORES DEGERATION factor of pollen wall (AMS) RETARDATION formation by (TDR) directly regulating biosynthetic pathways A crucial component of a DYSFUNCTIONAL Rice: Undeveloped bHLH transcription genetic network Preferentially in TAPETUM 1 Tapetum1 (UDT1) [S50, S51, S52] factor that controls anther tapetal cells (DYT1) Tomato: MS1035 development and function
Promotes aspartic Rice: ETERNAL proteases to TAPETUM 1 bHLH transcription trigger plant Tapetum [S53, S54] (EAT1)/ factor programmed cell bHLH141/DTD death
Promotes tapetal PCD and Rice: TDR Endothecium, bHLH transcription degeneration of INTERACTING middle layer, and [S55, S56] factor callose PROTEIN2 (TIP2) tapetum surrounding the microspores
Defective in Play a vital role in Highly expressed Tapetal MYB transcription tapetal in the tapetum, [S57] Development and factor differentiation and meiocytes and Function 1 (TDF1) function microspores
Involved in tapetum and pollen development and Tapetum, middle AtMYB103/MYB80 MYB transcription is required for the layers and [S58, S59, S60, /MS188 factor regulation of developing S61] tapetal microspores programmed cell death
Functions as a GA Tapetum and faint MYB transcription signaling factor in in anther wall Rice: GAMYB [S62, S63] factor anther layers and development microspore
Controls lipid bZIP transcription AtbZIP34 metabolism and/or Anthers and pistils [S64] factor cellular transport
Modify the Rice: transcription of PERSISTENT tapetal-specific TAPETAL CELL1 genes implicated in Specifically in (PTC1) pollen wall MALE STERILITY PHD finger motif tapetum shortly Petunia: development and [S65, S66, S67] 1 (MS1) transcription factor after microspore TAPETUM control tapetal release from tetrad DEVELOPMENT development by ZINC FINGER directly regulating PROTEIN1 (TAZ1) tapetal PCD and breakdown.
CALLOSE Regulates callose DEFECTIVEMICR CCCH-type zinc Meiocytes and the metabolism during [S68] OSPORE1 finger protein tapetum microsporogenesis (CDM1)
Stems and buds Involved in miRNA (microsporocytes CCCH zinc finger maturation and AtTTP and tapetal cells), [S69] protein pollen wall pattern weak expression in formation roots and leaves
WRKY34 and WRKY Required for male Pollen-specific [S70] WRKY2 transcription factor gametogenesis
Other regulators TRANSPOSABLE AT-hook family Determines nexine Tapetum [S71] ELEMENT protein formation SILENCING VIA AT-HOOK (TEK) Essential for pollen AUXIN Microsporocytes wall patterning by RESPONSE Auxin response and modulating [S72] FACTOR17 factor microgametophyte primexine (ARF17) s formation
Affected the timing RESPIRATORY- of tapetal PCD and BURST OXIDASE NADPH oxidase resulted in aborted Tapetal layer [S73] HOMOLOG E male (RBOHE) gametophytes
Regulates pre- Preferentially CYCLIN- mRNA splicing of expressed in leaf DEPEDENT Cyclin-dependent CALLOSE and inflorescence [S74] KINASE G1 protein kinase SYNTHASE5 and weakly (CDKG1) (CalS5) and pollen expressed wall formation in the other tissues
Arabidopsis Highly expressed Shaggy-like Shaggy-like protein Essential for pollen in floral buds [S75] protein kinases β kinases development especially in floral (ASKβ) stage 13-16
SIDECAR Plays a role in the POLLEN/LATERA male gametophyte Preferentially L ORGAN LBD transcription function primarily expressed in [S76] BOUNDARIES factors at germ cell pollen DOMAIN 10 mitosis. (LBD10)
Rice: Plays a critical role in rice tapetum cell development anther F-box protein Tapetum [S77] development and pollen formation. F-box (OsADF)
Plays an essential Brassica Non-coding RNA role in pollen campestris: Pollen-specific [S78] gene development and BcMF1 male fertility
Supplementary references
S1 Dong, X., et al. (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42, 315-328
S2 Enns, L.C., et al. (2005) Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol Biol. 58, 333- 349
S3 Shi, X., et al. (2015) GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol. 56, 497-509
S4 Hird, D.L., et al. (1993) The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to β-1, 3-glucanases. Plant J. 4, 1023-1033
S5 Wan, L., et al. (2011) A rice β-1, 3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta. 233, 309-323 S6 Paxson-Sowders, D.M., et al. (2001) DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 127, 1739- 1749
S7 Ariizumi, T., et al. (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39, 170-181
S8 Guan, Y.F., et al. (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 147, 852-863
S9 Sun, M.X., et al. (2013) Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 26, 83-91
S10 Chang, H.S., et al. (2012) No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiol. 158, 264-272
S11 Hu, J., et al. (2014) The Arabidopsis Exine Formation Defect (EFD) gene is required for primexine patterning and is critical for pollen fertility. New Phytol. 203, 140-154
S12 Yang, J., et al. (2013) Co-regulation of exine wall patterning, pollen fertility and anther dehiscence by Arabidopsis apyrases 6 and 7. Plant Physiol Biochem 69: 62-73
S13 Hsiao, A.-S., et al. (2014) The Arabidopsis cytosolic acyl-coA-binding proteins play combinatory roles in pollen development. Plant Cell Physiol. 56, 322-333
S14 Sumiyoshi, M., et al. (2014) UDP-Arabinopyranose mutase 3 is required for pollen wall morphogenesis in Rice (Oryza sativa). Plant Cell Physiol. 56, 232-241
S15 Chen, W., et al. (2011) Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 157, 842- 853
S16 Morant, M., et al. (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell. 19, 1473-1487
S17 Yang, X., et al. (2014) Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J Integr Plant Biol. 56, 979-994
S18 Dobritsa, A.A., et al. (2009) CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151, 574-589
S19 Li, H., et al. (2010) Cytochrome P450 family member CYP704B2 catalyzes the {omega}- hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell. 22, 173-190 S20 de Azevedo Souza, C., et al. (2009) A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell. 21, 507-525
S21 Kim, S.S., et al. (2010) LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl alpha-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell. 22, 4045-4066
S22 Grienenberger, E., et al. (2010) Analysis of TETRAKETIDE alpha-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell. 22, 4067-4083
S23 Aarts, M., et al. (1995) Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell. 7, 2115-2127
S24 Ariizumi, T., et al. (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol. 53, 107-116
S25 Haslam, T., et al. (2015) CER2-LIKE Proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol, 167, 682-692
S26 Jessen, D., et al. (2011) Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J. 68, 715-726
S27 Flores-Tornero, M., et al. (2015) Lack of phosphoserine phosphatase activity alters pollen and tapetum development in Arabidopsis thaliana. Plant Sci. 235, 81-88.
S28 Li, J., et al. (2010) The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 64, 482-497
S29 Schnurr, J.A., et al. (2006) UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta. 224, 520-532
S30 Moon, S., et al. (2013) Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol. 161, 663-675
S31 Ueda, K., et al. (2013) COLLAPSED ABNORMAL POLLEN1 gene encoding the arabinokinase-like protein is involved in pollen development in rice. Plant Physiol. 162, 858-871
S32 Jiang, J., et al. (2014) PECTATE LYASE-LIKE10 is associated with pollen wall development in Brassica campestris. J Integr Plant Biol. 56, 1095-1105
S33 Jiang, J., et al. (2014) PECTATE LYASE-LIKE 9 from Brassica campestris is associated with intine formation. Plant Sci. 229, 66-75
S34 Lin, S., et al. (2014) BcMF8, a putative arabinogalactan protein-encoding gene, contributes to pollen wall development, aperture formation and pollen tube growth in Brassica campestris. Ann Bot. 113,777-788 S35 Lyu, M., et al. (2015) BcMF26a and BcMF26b Are Duplicated Polygalacturonase Genes with Divergent Expression Patterns and Functions in Pollen Development and Pollen Tube Formation in Brassica campestris. PLoS One. doi:10.1371/journal.pone.0131173
S36 Lyu, M., et al. (2015) Identification and expression analysis of BoMF25, a novel polygalacturonase gene involved in pollen development of Brassica oleracea. Plant reproduction, 28, 121-132.
S37 Choi, H., et al. (2011) An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J. 65, 181-193
S38 Quilichini, T.D., et al. (2014) ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in Arabidopsis. Plant Cell. 26, 4483-4498
S39 Choi, H., et al. (2014) The role of Arabidopsis ABCG9 and ABCG31 ATP Binding Cassette Transporters in pollen fitness and the deposition of steryl glycosides on the pollen coat. Plant Cell. 26, 310-324
S40 Yadav, V., et al. (2014) ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell. 26, 3569-3588
S41 Zhu, L., et al. (2013) Post-meiotic deficient anther1 (PDA1) encodes an ABC transporter required for the development of anther cuticle and pollen exine in rice. J Plant Biol. 56, 59-68
S42 Qin, P., et al. (2013) ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in Rice. Plant Cell Physiol. 54, 138-154
S43 Huang, M.-D., et al. (2013) Abundant type III lipid transfer proteins in Arabidopsis tapetum are secreted to the locule and become a constituent of the pollen exine. Plant Physiol. 163, 1218-1229
S44 Zhang, D., et al. (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154, 149-162
S45 Li, J., et al. (2015) An endoplasmic reticulum magnesium transporter is essential for pollen development in Arabidopsis. Plant Sci. 231, 212-220
S46 Niemann, M.C., et al. (2015) Arabidopsis ROCK1 transports UDP-GlcNAc/UDP-GalNAc and regulates ER protein quality control and cytokinin activity. Proc Nat Acad Sci. 112, 291-296
S47 Xu, J., et al. (2014) ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell. 26, 1544-1556
S48 Li, N., et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 18, 2999-3014 S49 Xu, J., et al. (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell. 22, 91-107
S50 Zhang, W., et al. (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 133, 3085-3095
S51 Jeong, H.-J., et al. (2014) Tomato Male sterile 1035 is essential for pollen development and meiosis in anthers. J Exp Bot. 65, 6693-6709
S52 Jung, K.-H., et al. (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 17, 2705-2722
S53 Niu, N., et al. (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nature Commun. 4, 1445
S54 Ji, C., et al. (2013) A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol Plant. 6, 1715-1718
S55 Fu, Z., et al. (2014) The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell. 26, 1512-1524
S56 Ko, S.-S., et al. (2014) The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell. 26, 2486-2504
S57 Zhu, J., et al. (2008) Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 55, 266-277
S58 Zhang, Z.B., et al. (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 52, 528-538
S59 Phan, H.A., et al. (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell. 23, 2209-2224
S60 Phan, H.A., et al. (2012) MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol Boil. 78, 171-183
S61 Xu, Y., et al. (2014) MYB80 homologues in Arabidopsis, cotton and Brassica: regulation and functional conservation in tapetal and pollen development. BMC Plant Biol. 14, 278
S62 Aya, K., et al. (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell. 21, 1453-1472
S63 Aya, K., et al. (2011) The Gibberellin perception system evolved to regulate a pre- existing GAMYB-mediated system during land plant evolution. Nature Commun. 2, 544 S64 Gibalová, A., et al. (2009) AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Mol Biol. 70, 581- 601
S65 Yang, C., et al. (2007) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell. 19, 3530-3548
S66 Li, H., et al. (2011) PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156, 615-630
S67 Kapoor, S., et al. (2002) Silencing of the tapetum-specific zinc finger gene TAZ1 causes premature degeneration of tapetum and pollen abortion in petunia. Plant Cell. 14, 2353- 2367
S68 Lu, P., et al. (2014) The Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 gene is required for male fertility through regulating callose metabolism during microsporogenesis. Plant Physiol. 164, 1893-1904
S69 Shi, Z.-H., et al. (2015) Overexpression of AtTTP affects ARF17 expression and leads to male sterility in Arabidopsis. PLoS One. 10, e0117317
S70 Guan, Y., et al. (2014) Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 10, e1004384
S71 Lou, Y., et al. (2014) The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nature Commun. 5, 3855
S72 Yang, J., et al. (2013) AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiol. 162, 720-731
S73 Xie, H.-T., et al. (2014) Spatiotemporal production of reactive oxygen species by NADPH Oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. Plant Cell 26, 2007-2023
S74 Huang, X.Y., et al. (2013) CYCLIN-DEPENDENT KINASE G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis. Plant Cell. 25, 637-648
S75 Dong, X., et al. (2015) Suppression of ASKβ (AtSK32), a Clade III Arabidopsis GSK3, Leads to the Pollen Defect during Late Pollen Development. Molecules and cells. 38, 506-517
S76 Kim, M.J., et al. (2015) LATERAL ORGAN BOUNDARIES DOMAIN (LBD) 10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. The Plant Journal, 81, 794-809.
S77 Li, L., et al. (2015) An anther development F-box (ADF) protein regulated by tapetum degeneration retardation (TDR) controls rice anther development. Planta. 241, 157-166
S78 Song, J.-H., et al. (2013) BcMF11, a novel non-coding RNA gene from Brassica campestris, is required for pollen development and male fertility. Plant Cell Rep. 32, 21-30