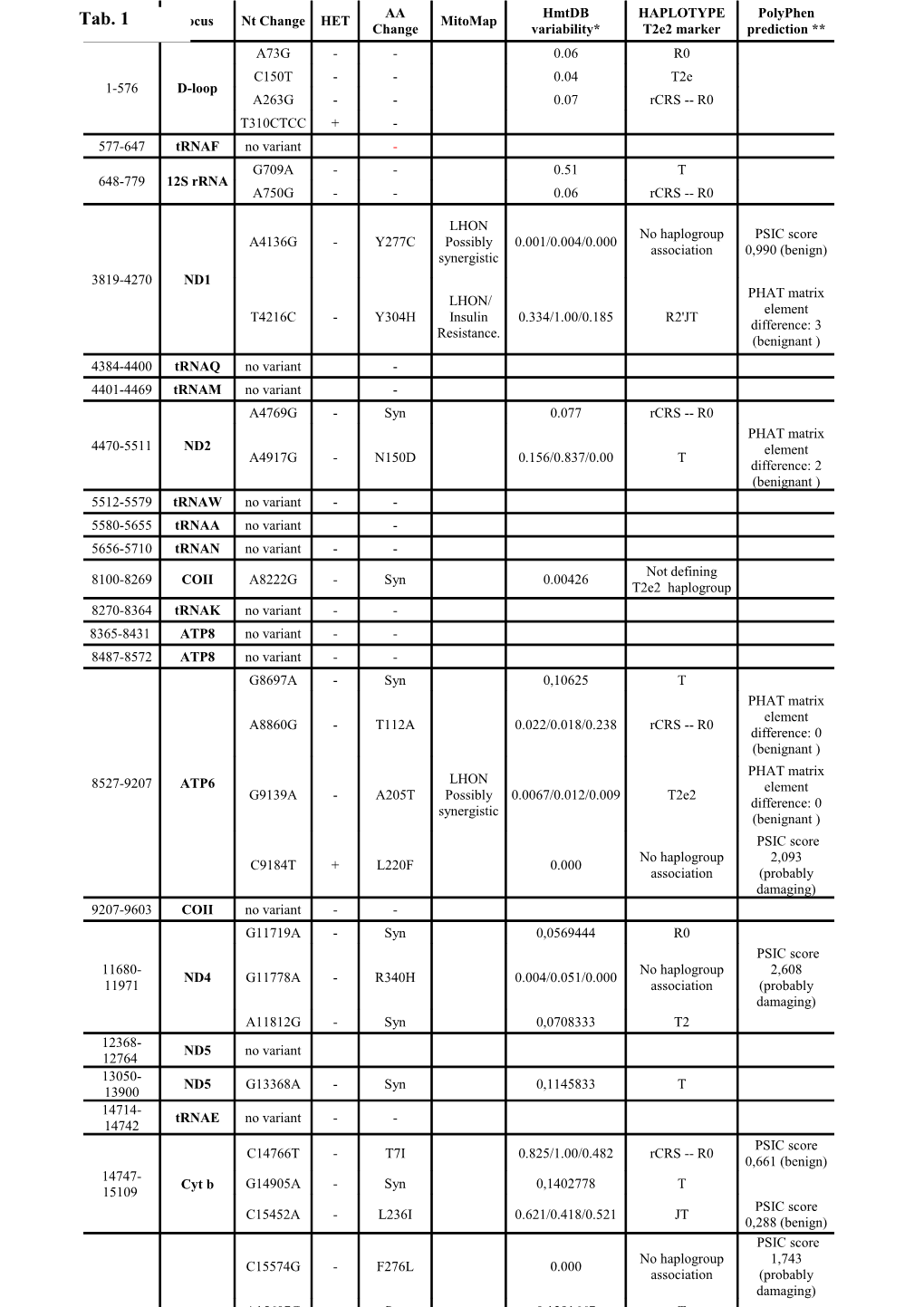
AA HmtDB HAPLOTYPE PolyPhen Tab. 1 Locus Nt Change HET MitoMap Change variability* T2e2 marker prediction ** A73G - - 0.06 R0 C150T - - 0.04 T2e 1-576 D-loop A263G - - 0.07 rCRS -- R0 T310CTCC + - 577-647 tRNAF no variant - G709A - - 0.51 T 648-779 12S rRNA A750G - - 0.06 rCRS -- R0
LHON No haplogroup PSIC score A4136G - Y277C Possibly 0.001/0.004/0.000 association 0,990 (benign) synergistic 3819-4270 ND1 PHAT matrix LHON/ element T4216C - Y304H Insulin 0.334/1.00/0.185 R2'JT difference: 3 Resistance. (benignant ) 4384-4400 tRNAQ no variant - 4401-4469 tRNAM no variant - A4769G - Syn 0.077 rCRS -- R0 PHAT matrix 4470-5511 ND2 element A4917G - N150D 0.156/0.837/0.00 T difference: 2 (benignant ) 5512-5579 tRNAW no variant - - 5580-5655 tRNAA no variant - 5656-5710 tRNAN no variant - - Not defining 8100-8269 COII A8222G - Syn 0.00426 T2e2 haplogroup 8270-8364 tRNAK no variant - - 8365-8431 ATP8 no variant - - 8487-8572 ATP8 no variant - - G8697A - Syn 0,10625 T PHAT matrix element A8860G - T112A 0.022/0.018/0.238 rCRS -- R0 difference: 0 (benignant ) PHAT matrix LHON 8527-9207 ATP6 element G9139A - A205T Possibly 0.0067/0.012/0.009 T2e2 difference: 0 synergistic (benignant ) PSIC score No haplogroup 2,093 C9184T + L220F 0.000 association (probably damaging) 9207-9603 COII no variant - - G11719A - Syn 0,0569444 R0 PSIC score 11680- No haplogroup 2,608 ND4 G11778A - R340H 0.004/0.051/0.000 11971 association (probably damaging) A11812G - Syn 0,0708333 T2 12368- ND5 no variant 12764 13050- ND5 G13368A - Syn 0,1145833 T 13900 14714- tRNAE no variant - - 14742 PSIC score C14766T - T7I 0.825/1.00/0.482 rCRS -- R0 0,661 (benign) 14747- Cyt b G14905A - Syn 0,1402778 T 15109 PSIC score C15452A - L236I 0.621/0.418/0.521 JT 0,288 (benign) PSIC score No haplogroup 1,743 C15574G - F276L 0.000 association (probably damaging) A15607G - Syn 0,1291667 T * For non synonimous changes three values are reported: human nucleotide (nt) variability/human aminoacidic (aa) variability/inter-mammalian aa variability. Variability is calculated according to [1]). ** For a substitution in an annotated or predicted transmembrane region, PolyPhen uses the PHAT transmembrane specific matrix score to evaluate possible functional effect of a nsSNP in the transmembrane region. IF PHAT is negative the variant is damaging when the variant is in a transmembrane region. In non transmembrane region the PSIC score is estimated: higher the PSIC value higher is the damage caused by the mutation.
Direct sequencing of almost the entire molecules of mtDNA from the patient was carried out with a 310 Sequence Analyzer. Tab. 1 reports the sequenced fragments and the 32 variant sites: 23 of this sites allow the association of the studied mtDNA with T2e2 haplogroup; this is defined by 31 sites, but eight sites are undetermined because not sequenced. Besides the 11778/ND4 LHON primary mutation, the genome harboured nine non-synonymous nucleotide changes of which six (4216, 4917, 8860, 9139, 14766 and 15452) contributing to the definition of T2e2 haplogroup and his roots (according to PhyloTree annotations [http://www.phylotree.org]) [2]. Checking through PolyPhen [http://genetics.bwh.harvard.edu/pph/] the effect of the mutation on both the structure and/or the function of the protein, the above six variants results benignant, according to the calculated scores (see notes in Tab. 1). Data in HmtDB [3] confirm this hypothesis thanks to their variability estimated on about 7000 complete human mitochondrial genomes: significant variability values show the presence of this mutation in several genomes thus it can be affirmed that these variants are positively selected in the population. The remaining three of the non-synonymous nucleotide changes are all not associated to haplogroups: the A4136G (T>C in ND1) is present in different populations and reports a not relevant PSIC score; on the contrary, C15574G and C15608G, are not associated to haplogroups, not reported both in HmtDB and in MitoMAP and show a high PSIC value indicative of a damaging effect on protein function. A careful comparison of the mtDNA sequence of the presented patient with that of family 1 with LHON plus myoclonus described in the paper by La Morgia et al. [4] disclosed that they both harbour the same non-synonymous changes such as the A4136G and the T4216C ND1 mutations, the A4917G ND2 mutation and the G9139A ATPase6 mutation (Tab. 1), suggesting that they come from the same geographic area. However, our patient harbours two further non synonymous Cytb mutations (C15574G and C15608G) whose statistics reported in Tab. 1 indicate a potentially damaging role. Therefore, we can conclude that it is high the probability that the case herein presented is possibly related to a cluster of LHON cases living in Southern Italy and that all belong to a common female founder as the family 1 in [4] but surely he should not be a component of the same family.
References [1] Accetturo M, Santamaria M, Lascaro D, Rubino F, Achilli A, Torroni A, Tommaseo-Ponzetta M, Attimonelli M. (2006) Human mtDNA site-specific variability values can act as haplogroup markers. Hum Mutat. 27:965-74. [2] van Oven M, Kayser M. (2009) Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation.Hum Mutat.; 30(2):E386-E394. [3] Attimonelli M, Accetturo M, Santamaria M, Lascaro D, Scioscia G, Pappadà G, Russo L, Zanchetta L, Tommaseo-Ponzetta M. (2005) HmtDB, a human mitochondrial genomic resource based on variability studies supporting population genetics and biomedical research. BMC Bioinformatics.;6 , Suppl 4:S4. [4] La Morgia C, Achilli A, Iommarini L, Barboni P, Pala M, Olivieri A, Zanna C, Vidoni S, Tonon C, Lodi R, Vetrugno R, Mostacci B, Liguori R, Carroccia R, Montagna P, Rugolo M, Torroni A, Carelli V. (2008) Rare mtDNA variants in Leber hereditary optic neuropathy families with recurrence of myoclonus. Neurology.; 70(10):762-70.