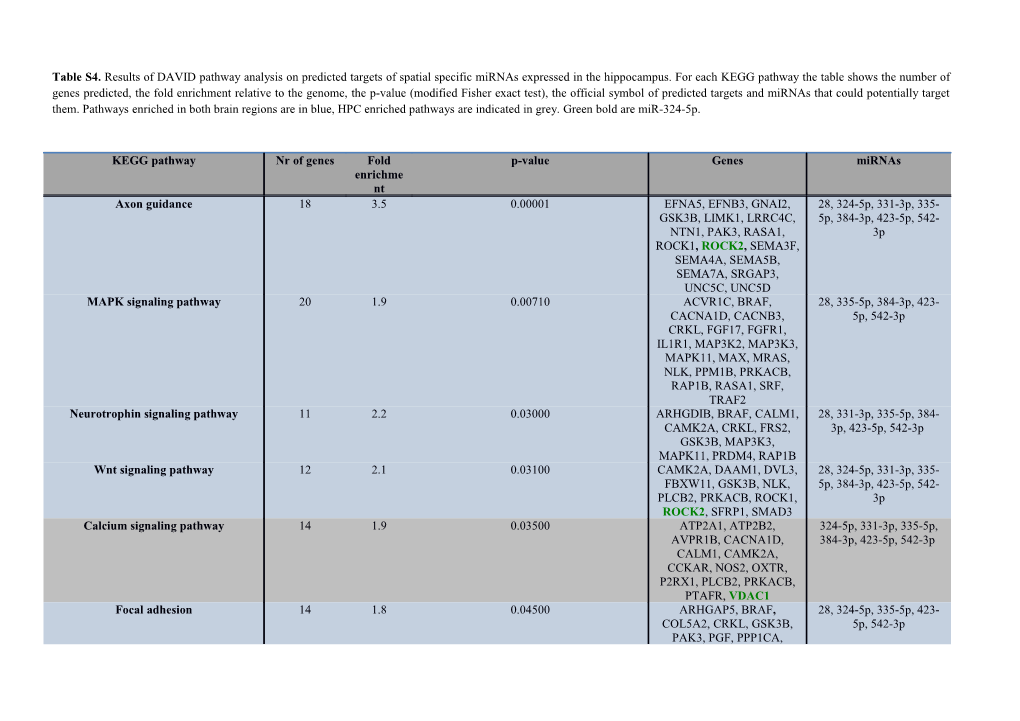
Table S4. Results of DAVID pathway analysis on predicted targets of spatial specific miRNAs expressed in the hippocampus. For each KEGG pathway the table shows the number of genes predicted, the fold enrichment relative to the genome, the p-value (modified Fisher exact test), the official symbol of predicted targets and miRNAs that could potentially target them. Pathways enriched in both brain regions are in blue, HPC enriched pathways are indicated in grey. Green bold are miR-324-5p.
KEGG pathway Nr of genes Fold p-value Genes miRNAs enrichme nt Axon guidance 18 3.5 0.00001 EFNA5, EFNB3, GNAI2, 28, 324-5p, 331-3p, 335- GSK3B, LIMK1, LRRC4C, 5p, 384-3p, 423-5p, 542- NTN1, PAK3, RASA1, 3p ROCK1, ROCK2, SEMA3F, SEMA4A, SEMA5B, SEMA7A, SRGAP3, UNC5C, UNC5D MAPK signaling pathway 20 1.9 0.00710 ACVR1C, BRAF, 28, 335-5p, 384-3p, 423- CACNA1D, CACNB3, 5p, 542-3p CRKL, FGF17, FGFR1, IL1R1, MAP3K2, MAP3K3, MAPK11, MAX, MRAS, NLK, PPM1B, PRKACB, RAP1B, RASA1, SRF, TRAF2 Neurotrophin signaling pathway 11 2.2 0.03000 ARHGDIB, BRAF, CALM1, 28, 331-3p, 335-5p, 384- CAMK2A, CRKL, FRS2, 3p, 423-5p, 542-3p GSK3B, MAP3K3, MAPK11, PRDM4, RAP1B Wnt signaling pathway 12 2.1 0.03100 CAMK2A, DAAM1, DVL3, 28, 324-5p, 331-3p, 335- FBXW11, GSK3B, NLK, 5p, 384-3p, 423-5p, 542- PLCB2, PRKACB, ROCK1, 3p ROCK2, SFRP1, SMAD3 Calcium signaling pathway 14 1.9 0.03500 ATP2A1, ATP2B2, 324-5p, 331-3p, 335-5p, AVPR1B, CACNA1D, 384-3p, 423-5p, 542-3p CALM1, CAMK2A, CCKAR, NOS2, OXTR, P2RX1, PLCB2, PRKACB, PTAFR, VDAC1 Focal adhesion 14 1.8 0.04500 ARHGAP5, BRAF, 28, 324-5p, 335-5p, 423- COL5A2, CRKL, GSK3B, 5p, 542-3p PAK3, PGF, PPP1CA, PTEN, RAP1B, ROCK1, ROCK2, THBS3, TLN2 Long-term potentiation 7 2.6 0.05500 BRAF, CALM1, CAMK2A, 28, 331-3p, 335-5p, 384- PLCB2, PPP1CA, PRKACB, 3p, 423-5p RAP1B Taurine and hypotaurine metabolism 3 7.7 0.05600 CSAD, GAD2, GGT7 324-5p, 384-3p, 542-3p Hedgehog signaling pathway 6 2.8 0.05800 BMP7, CSNK1D, 28, 331-3p, 335-5p, 384- CSNK1G1, FBXW11, 3p, 542-3p GSK3B, PRKACB