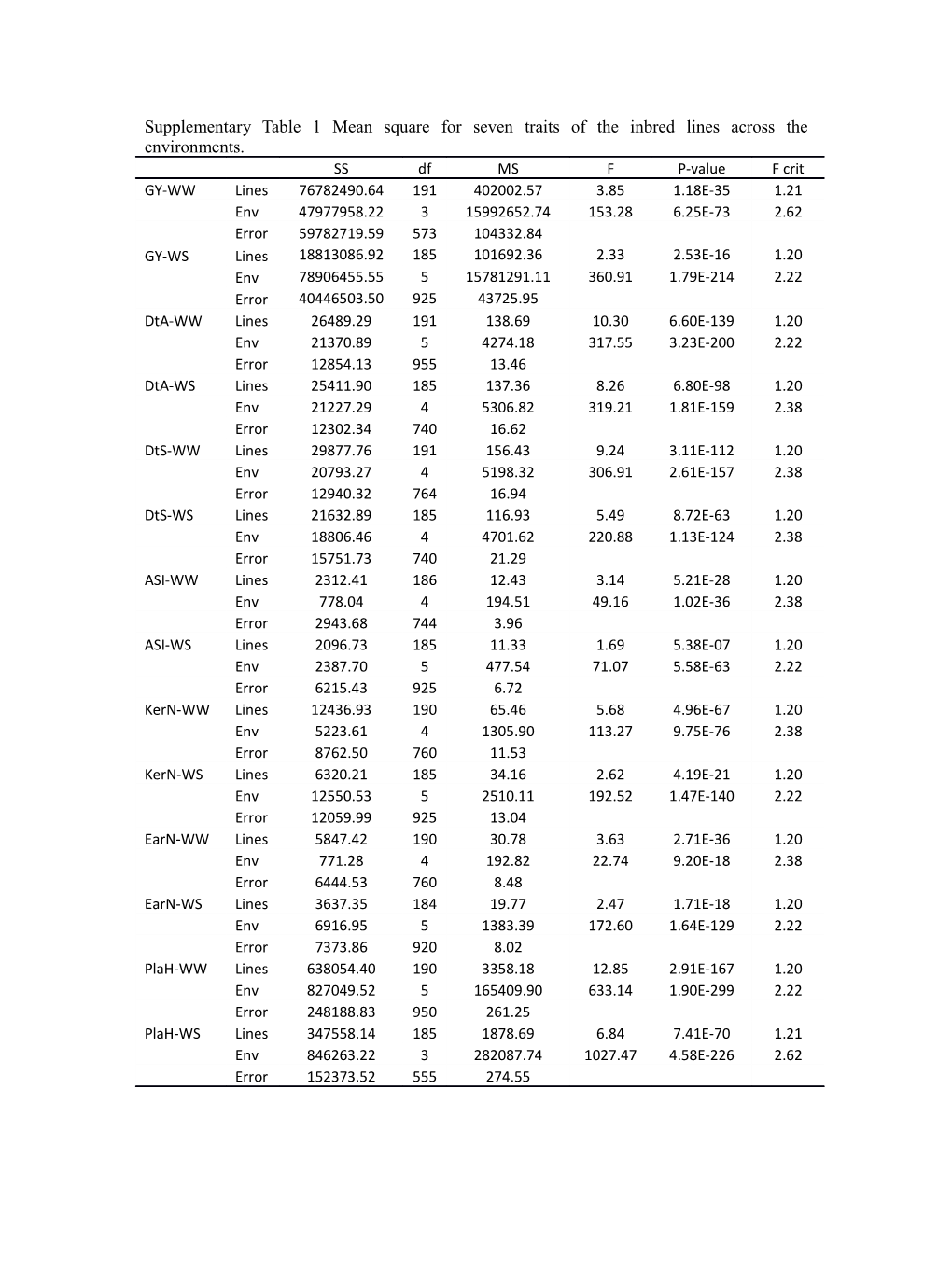
Supplementary Table 1 Mean square for seven traits of the inbred lines across the environments. SS df MS F P-value F crit GY-WW Lines 76782490.64 191 402002.57 3.85 1.18E-35 1.21 Env 47977958.22 3 15992652.74 153.28 6.25E-73 2.62 Error 59782719.59 573 104332.84 GY-WS Lines 18813086.92 185 101692.36 2.33 2.53E-16 1.20 Env 78906455.55 5 15781291.11 360.91 1.79E-214 2.22 Error 40446503.50 925 43725.95 DtA-WW Lines 26489.29 191 138.69 10.30 6.60E-139 1.20 Env 21370.89 5 4274.18 317.55 3.23E-200 2.22 Error 12854.13 955 13.46 DtA-WS Lines 25411.90 185 137.36 8.26 6.80E-98 1.20 Env 21227.29 4 5306.82 319.21 1.81E-159 2.38 Error 12302.34 740 16.62 DtS-WW Lines 29877.76 191 156.43 9.24 3.11E-112 1.20 Env 20793.27 4 5198.32 306.91 2.61E-157 2.38 Error 12940.32 764 16.94 DtS-WS Lines 21632.89 185 116.93 5.49 8.72E-63 1.20 Env 18806.46 4 4701.62 220.88 1.13E-124 2.38 Error 15751.73 740 21.29 ASI-WW Lines 2312.41 186 12.43 3.14 5.21E-28 1.20 Env 778.04 4 194.51 49.16 1.02E-36 2.38 Error 2943.68 744 3.96 ASI-WS Lines 2096.73 185 11.33 1.69 5.38E-07 1.20 Env 2387.70 5 477.54 71.07 5.58E-63 2.22 Error 6215.43 925 6.72 KerN-WW Lines 12436.93 190 65.46 5.68 4.96E-67 1.20 Env 5223.61 4 1305.90 113.27 9.75E-76 2.38 Error 8762.50 760 11.53 KerN-WS Lines 6320.21 185 34.16 2.62 4.19E-21 1.20 Env 12550.53 5 2510.11 192.52 1.47E-140 2.22 Error 12059.99 925 13.04 EarN-WW Lines 5847.42 190 30.78 3.63 2.71E-36 1.20 Env 771.28 4 192.82 22.74 9.20E-18 2.38 Error 6444.53 760 8.48 EarN-WS Lines 3637.35 184 19.77 2.47 1.71E-18 1.20 Env 6916.95 5 1383.39 172.60 1.64E-129 2.22 Error 7373.86 920 8.02 PlaH-WW Lines 638054.40 190 3358.18 12.85 2.91E-167 1.20 Env 827049.52 5 165409.90 633.14 1.90E-299 2.22 Error 248188.83 950 261.25 PlaH-WS Lines 347558.14 185 1878.69 6.84 7.41E-70 1.21 Env 846263.22 3 282087.74 1027.47 4.58E-226 2.62 Error 152373.52 555 274.55 Supplementary Table 2 Association signals information for seven traits related to drought tolerance in seven environments Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
08XJ GRMZM2G17465 DtS-WS SYN9378 1 3.34E+06 8.97E-05 0.09 promoter VQ 1 0_P01
GRMZM2G36874 DtA-WW 07XJ SYN6207 1 8.92E+06 9.17E-05 0.11 promoter unknown 4_P01
AC194905.2_FGT KerN-WS 06HN SYN3029 1 7.23E+07 5.51E-06 0.17 promoter unknown 001
ER lumen 08XJ GRMZM2G03765 protein DtS-WS PZE-101101218 1 9.70E+07 7.09E-05 0.09 intron 1 5_P01 retaining receptor
GRMZM2G05514 KerN-WS 06HN PZE-101108052 1 1.13E+08 9.63E-05 0.09 intron unknown 3_P01
GRMZM2G58426 ASI-WS 09XJ PZE-101120408 1 1.48E+08 7.11E-05 0.10 promoter unknown 0_P01
APOBEC/CMP GRMZM2G03664 PlaH-WW 07HN SYN35039 1 1.92E+08 1.31E-05 0.14 3 UTR deaminase, 0_P02 zinc-binding
Helix-loop- GRMZM2G01980 PlaH-WW 07HN SYN35041 1 1.92E+08 8.38E-05 0.09 5 UTR helix DNA- 6_P01 binding Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
GRMZM2G56185 DtS-WW 07XJ PZE-101163883 1 2.07E+08 5.05E-05 0.11 promoter unknown 4_P01
GRMZM2G16693 FAD ASI-WW 07XJ PZE-101167516 1 2.10E+08 9.25E-05 0.11 3 UTR 1_P01 synthetase
Cytochrome GRMZM2G47044 ASI-WS 07XJ PZE-101180354 1 2.24E+08 5.75E-05 0.12 Exon P450, E-class, 2_P03 group I
Peptidyl-prolyl GRMZM2G09658 cis-trans ASI-WS 09XJ SYN37125 1 2.62E+08 8.80E-05 0.09 5 UTR 5_P06 isomerase, FKBP-type
Aminoacyl- GRMZM2G09680 tRNAsyntheta ASI-WS 09XJ SYN37124 1 2.62E+08 2.84E-05 0.10 Exon 6_P01 se, class I, conserved site
Protein of GRMZM2G09469 unknown ASI-WW 07XJ PZE-101214133 1 2.64E+08 5.58E-06 0.17 3 UTR 8_P01 function DUF1336
Protein of GRMZM2G09469 unknown KerN-WW 09XJ PZE-101219533 1 2.70E+08 5.86E-06 0.17 3 UTR 8_P01 function DUF1336 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
GRMZM2G04968 PlaH-WW 09XJ SYN21892 1 2.88E+08 4.85E-05 0.12 3 UTR unknown 7_P01
Heat shock GRMZM2G11545 factor (HSF)- PlaH-WW 09XJ SYN37548 1 2.88E+08 2.59E-05 0.10 Exon 6_P01 type, DNA- binding
08XJ GRMZM2G04688 Pentatricopep PlaH-WW SYN38957 1 2.98E+08 8.45E-05 0.09 promoter 1 8_P01 tide repeat
08XJ GRMZM2G42680 ASI-WW PZE-101257312 1 3.00E+08 6.24E-05 0.10 5 UTR WD40 repeat 1 2_P01
08XJ GRMZM2G12926 Zinc finger, GY-WS SYN27290 2 1.70E+07 5.44E-06 0.27 Exon 2 1_P03 C2H2-like
08XJ AC205661.3_FGT ASI-WW PZE-102085765 2 7.66E+07 3.31E-05 0.13 intron unknown 2 004
08XJ DtS-WW PZE-102101935 2 1.21E+08 2.25E-05 0.12 NA 1
AC190936.1_FGT Antifreeze EarN-WS 06HN PZA02939.5 2 1.57E+08 9.00E-05 0.09 promoter 009 protein, type I
GRMZM2G40016 EarN-WW 07XJ PZE-102159259 2 2.03E+08 8.72E-05 0.09 3 UTR unknown 9_P01 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Alpha-L- arabinofurano GRMZM2G09727 ASI-WS 06HN PZA02471.5 2 2.15E+08 1.67E-06 0.14 intron sidase, C- 7_P01 terminalC- terminal
GRMZM2G15584 Cyclin-like F- DtS-WW 08HN SYN18368 2 2.20E+08 8.68E-05 0.09 Exon 9_P01 box
GRMZM2G15584 Cyclin-like F- DtS-WW 08HN SYN18369 2 2.20E+08 8.68E-05 0.09 3 UTR 9_P01 box
Peptidase S8 and S53, 08XJ GRMZM2G34562 KerN-WW PZE-102184023 2 2.24E+08 4.90E-05 0.09 Exon subtilisin, 2 2_P01 kexin, sedolisin
PUT-163a- GRMZM2G09795 KerN-WW 07HN 3 2.41E+06 5.90E-05 0.11 3 UTR unknown 74244282-3684 9_P01
GRMZM2G70130 DtS-WS 09XJ PZE-103015618 3 8.41E+06 9.61E-05 0.11 promoter unknown 1_P01
DNA mismatch GRMZM2G11021 PlaH-WW 07HN PZE-103016554 3 9.29E+06 9.13E-05 0.09 intron repair protein 2_P01 MutS, C- terminal Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
ASI-WS 09XJ PZE-103070836 3 1.12E+08 1.46E-05 0.14 NA
ASI-WS 09XJ PZE-103071028 3 1.13E+08 1.74E-05 0.13 NA
08XJ ASI-WS PZE-103072939 3 1.17E+08 4.98E-05 0.14 NA 2
ASI-WS 09XJ PZE-103087762 3 1.42E+08 3.30E-05 0.12 NA
Concanavalin GRMZM2G17424 A-like GY-WW 07XJ PZE-103087821 3 1.42E+08 8.75E-05 0.09 intron 9_P03 lectin/glucana se
Concanavalin GRMZM2G17424 A-like GY-WW 07XJ PZE-103087822 3 1.42E+08 2.40E-05 0.10 intron 9_P03 lectin/glucana se
GRMZM2G30291 DtA-WW 07XJ SYN32914 3 1.43E+08 9.90E-05 0.09 Exon unknown 3_P01
Amino GRMZM2G03081 ASI-WS 09XJ PZE-103091618 3 1.48E+08 1.39E-06 0.17 Exon acid/polyamin 4_P01 e transporter I
GRMZM2G49506 DtA-WW 07XJ PZE-103091926 3 1.48E+08 4.05E-05 0.10 promoter unknown 5_P01 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Ribosomal GRMZM2G08110 EarN-WW 07XJ PZE-103098294 3 1.57E+08 9.08E-05 0.09 5 UTR protein L13e, 2_P02 conserved site
GRMZM2G05629 Tetratricopept DtA-WS 08HN SYN31293 3 1.64E+08 7.33E-05 0.08 3 UTR 6_P01 ide TPR2
GRMZM2G32906 DtA-WW 07XJ PZE-103108797 3 1.68E+08 4.68E-06 0.12 promoter unknown 9_P01
GRMZM2G32906 DtS-WW 07XJ PZE-103108797 3 1.68E+08 8.12E-05 0.09 promoter unknown 9_P01
08XJ GRMZM2G32906 DtS-WW PZE-103108797 3 1.68E+08 4.29E-05 0.10 promoter unknown 2 9_P01
DtA-WW 07XJ PZE-103108908 3 1.68E+08 9.84E-06 0.11 NA
08XJ DtA-WW PZE-103108908 3 1.68E+08 3.10E-05 0.10 NA 2
EarN-WW 07HN PZE-103122953 3 1.79E+08 9.82E-05 0.09 NA
Alpha/beta GRMZM2G12608 EarN-WS 07XJ PZE-103124398 3 1.80E+08 4.51E-05 0.10 Exon hydrolase 3_P01 fold-1
Sodium:dicarb 08XJ GRMZM2G31576 DtA-WW PZE-103126852 3 1.82E+08 2.90E-05 0.10 intron oxylate 1 9_P01 symporter Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Sodium:dicarb GRMZM2G31576 DtA-WS 07XJ PZE-103126852 3 1.82E+08 6.79E-05 0.09 intron oxylate 9_P01 symporter
Sodium:dicarb 08XJ GRMZM2G31576 DtA-WS PZE-103126852 3 1.82E+08 7.88E-06 0.12 intron oxylate 1 9_P01 symporter
Sodium:dicarb 08XJ GRMZM2G31576 DtA-WW PZE-103126857 3 1.82E+08 2.90E-05 0.10 5 UTR oxylate 1 9_P01 symporter
Sodium:dicarb GRMZM2G31576 DtA-WS 07XJ PZE-103126857 3 1.82E+08 6.79E-05 0.09 5 UTR oxylate 9_P01 symporter
Sodium:dicarb 08XJ GRMZM2G31576 DtA-WS PZE-103126857 3 1.82E+08 7.88E-06 0.12 5 UTR oxylate 1 9_P01 symporter
GRMZM2G70203 KerN-WS 06HN SYN9245 3 1.82E+08 8.37E-05 0.10 3 UTR unknown 6_P01
08XJ DtA-WS PZE-103127059 3 1.82E+08 5.08E-06 0.13 NA 1
Purple acid GRMZM2G09310 phosphatase- GY-WW 07HN SYN6034 3 1.90E+08 1.99E-05 0.13 3 UTR 1_P01 like, N- terminal Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
08XJ DtS-WS PZE-103169160 3 2.16E+08 9.51E-05 0.09 NA 1
AC184699.3_FGT KerN-WS 09XJ PZE-103174030 3 2.19E+08 4.73E-05 0.13 Exon unknown 005
08XJ GRMZM2G00299 Zinc finger, DtS-WW PZE-103184446 3 2.28E+08 7.18E-05 0.11 Exon 1 9_P01 MIZ-type
08XJ GRMZM2G40760 Antifreeze DtA-WW PZE-104012524 4 1.08E+07 6.75E-05 0.11 promoter 1 5_P01 protein, type I
PlaH-WS 06HN PZE-104016012 4 1.56E+07 7.00E-05 0.15 NA
Myeloid transforming GRMZM2G01354 PlaH-WS 06HN SYN30703 4 1.74E+07 9.10E-05 0.12 Exon gene on 6_P01 chromosome 8 (MTG8)
U2 auxiliary GRMZM2G14651 PlaH-WS 06HN PZA00139.4 4 2.01E+07 1.62E-05 0.12 Exon factor small 4_P01 subunit
GRMZM2G10144 Antifreeze PlaH-WS 06HN PZE-104019775 4 2.04E+07 3.10E-05 0.11 intron 6_P01 protein, type I Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Protein of GRMZM2G17142 unknown PlaH-WS 06HN SYN24029 4 2.06E+07 7.28E-05 0.10 intron 0_P01 function DUF760
GRMZM2G15021 GY-WW 07XJ PZE-104020148 4 2.13E+07 4.25E-05 0.11 Exon unknown 5_P02
GRMZM2G15021 KerN-WW 07XJ PZE-104020148 4 2.13E+07 1.26E-05 0.13 Exon unknown 5_P02
GRMZM2G12295 PlaH-WS 06HN SYN23840 4 2.18E+07 8.00E-05 0.11 Exon unknown 4_P01
GRMZM2G09881 Mob1/phocei PlaH-WS 06HN PZE-104025828 4 3.08E+07 5.90E-05 0.11 intron 9_P02 n
GRMZM2G56615 ASI-WW 07HN PZE-104033229 4 4.12E+07 3.19E-05 0.10 promoter unknown 7_P01
EarN-WW 07XJ PZE-104049834 4 8.45E+07 9.67E-05 0.11 NA
08XJ GRMZM2G52512 DtA-WW PZE-104082002 4 1.56E+08 8.31E-06 0.11 promoter unknown 2 1_P01
GRMZM2G52512 DtA-WW 09XJ PZE-104082002 4 1.56E+08 1.85E-05 0.11 promoter unknown 1_P01
EarN-WW 07HN PZE-104097009 4 1.73E+08 7.77E-05 0.09 NA Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
08XJ GY-WS PZE-104123190 4 2.06E+08 8.17E-05 0.26 NA 2
Peptidase C19, ubiquitin 08XJ GRMZM2G11631 carboxyl- KerN-WS PZE-104139486 4 2.33E+08 1.57E-05 0.11 3 UTR 1 4_P01 terminal hydrolase 2, conserved site
08XJ GRMZM2G13937 DtS-WS PZE-104153023 4 2.44E+08 5.00E-05 0.12 5 UTR unknown 1 2_P04
Alpha-1,4- PUT-163a- GRMZM2G08732 glucan-protein PlaH-WS 07XJ 5 1.87E+07 8.70E-05 0.11 Exon 16925922-1125 6_P01 synthase, UDP-forming
Alpha-1,4- 08XJ PUT-163a- GRMZM2G08732 glucan-protein PlaH-WS 5 1.87E+07 3.74E-05 0.12 Exon 1 16925922-1125 6_P01 synthase, UDP-forming
Alpha-1,4- 08XJ PUT-163a- GRMZM2G08732 glucan-protein PlaH-WW 5 1.87E+07 3.61E-05 0.12 Exon 1 16925922-1125 6_P01 synthase, UDP-forming Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
08XJ KerN-WS PZE-105038171 5 2.29E+07 2.43E-05 0.23 NA 2
08XJ GRMZM2G02689 Zinc finger, ZZ- KerN-WS SYN2222 5 2.31E+07 5.16E-05 0.21 promoter 2 2_P04 type
Glycosyltransf GRMZM2G07489 erase AER61, ASI-WS 09XJ PZE-105066227 5 6.77E+07 2.62E-05 0.16 intron 6_P01 uncharacteriz ed
08XJ PUT-163a- GRMZM2G06219 DtA-WW 5 1.31E+08 8.21E-05 0.11 3 UTR unknown 2 149043264-814 9_P01
08XJ GRMZM2G05392 DtA-WW SYN32230 5 1.35E+08 8.79E-05 0.09 Exon VHS subgroup 2 5_P01
AC211190.4_FGT EarN-WS 07XJ PZE-105098019 5 1.44E+08 2.28E-05 0.10 intron unknown 001
ASI-WW 07XJ PZE-105100076 5 1.49E+08 3.76E-06 0.20 NA
08XJ ASI-WW PZE-105100076 5 1.49E+08 2.74E-05 0.15 NA 1
GRMZM2G09296 EarN-WS 07XJ PZE-105100386 5 1.50E+08 9.38E-05 0.09 promoter unknown 8_P01
Mercury 08XJ PUT-163a- GRMZM2G11333 DtA-WW 5 1.68E+08 1.59E-05 0.11 Exon scavenger 2 18177956-1457 2_P03 protein Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
GRMZM2G14416 EarN-WS 08HN SYN15524 5 1.80E+08 4.30E-05 0.10 5 UTR unknown 6_P01
08XJ GRMZM2G41382 Antifreeze DtS-WW PZE-105145239 5 1.98E+08 7.24E-05 0.11 promoter 1 9_P01 protein, type I
Target SNARE 08XJ GRMZM2G10157 ASI-WS SYN3700 5 2.15E+08 2.27E-05 0.12 intron coiled-coil 2 1_P02 region
Target SNARE 08XJ PUT-163a- GRMZM2G10157 ASI-WS 5 2.15E+08 2.27E-05 0.12 3 UTR coiled-coil 2 71328889-3180 1_P02 region
GRMZM2G12322 Synaptojanin, KerN-WS 06HN PZE-106001375 6 2.38E+06 2.42E-05 0.10 intron 7_P01 N-terminal
GRMZM2G34224 Harpin- DtA-WW 09XJ SYN11615 6 7.77E+07 9.03E-05 0.09 intron 3_P01 induced 1
GRMZM2G34224 Harpin- DtA-WW 09XJ SYN11613 6 7.77E+07 1.36E-05 0.12 Exon 3_P01 induced 1
08XJ GRMZM2G34224 Harpin- DtS-WS SYN11613 6 7.77E+07 6.71E-05 0.09 Exon 1 3_P01 induced 1
GRMZM2G01268 Protein EarN-WS 08HN PZE-106058073 6 1.09E+08 2.24E-05 0.13 Exon 5_P01 kinase-like Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Potassium channel, GRMZM2G15625 PlaH-WS 06HN PZE-106063999 6 1.16E+08 7.94E-05 0.12 Exon voltage- 5_P01 dependent, EAG/ELK/ER
08XJ GRMZM2G04633 GY-WS PZE-106079456 6 1.36E+08 7.55E-05 0.14 3 UTR unknown 1 1_P01
Ribosomal PUT-163a- GRMZM2G12857 RNA KerN-WS 07XJ 6 1.65E+08 7.93E-05 0.09 Exon 76287889-3910 9_P03 methyltransfe raseRrmJ/FtsJ
GRMZM2G13292 Ribosomal ASI-WW 09XJ SYN38602 6 1.66E+08 6.28E-05 0.11 promoter 9_P04 protein S12e
08XJ GRMZM2G42170 Tubby, N- DtA-WW SYN10743 7 1.30E+06 6.00E-05 0.09 promoter 2 7_P01 terminal
PUT-163a- GRMZM2G01323 PlaH-WS 06HN 7 5.15E+06 9.88E-05 0.10 Exon EF-HAND 1 71419466-3204 6_P02
Protein of GRMZM2G02958 unknown PlaH-WS 06HN PZE-107009409 7 6.03E+06 4.01E-06 0.14 3 UTR 7_P01 function DUF266, plant
GRMZM2G13674 ASI-WS 06HN PZE-107083353 7 1.32E+08 2.71E-05 0.13 promoter unknown 8_P01 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Alpha/beta 08XJ AC155434.2_FGT ASI-WW SYN37421 7 1.68E+08 4.82E-05 0.10 Exon hydrolase 2 001 fold-1
GRMZM2G00175 Peptidase S14, GY-WS 07HN PZE-108036539 8 5.29E+07 6.06E-05 0.15 intron 5_P01 ClpP
GY-WS 07HN PZE-108050917 8 8.79E+07 4.82E-05 0.15 NA
08XJ PUT-163a- GRMZM2G09318 DtS-WW 8 8.96E+07 5.33E-05 0.09 Exon RasGTPase 1 18163705-1263 6_P01
DtA-WW 07XJ PZE-108052412 8 9.18E+07 6.06E-05 0.09 NA
08XJ DtA-WW PZE-108052412 8 9.18E+07 3.84E-05 0.10 NA 1
DtS-WW 07XJ PZE-108052412 8 9.18E+07 2.35E-05 0.10 NA
08XJ DtS-WW PZE-108052412 8 9.18E+07 6.60E-05 0.09 NA 1
08XJ DtA-WW PZE-108052490 8 9.19E+07 8.33E-06 0.11 NA 1
Peptidyl-tRNA 08XJ GRMZM2G12784 DtA-WW PZE-108071041 8 1.23E+08 2.47E-05 0.10 promoter hydrolase, 1 4_P02 PTH2 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Peptidyl-tRNA 08XJ GRMZM2G12784 PlaH-WS PZE-108071041 8 1.23E+08 7.63E-05 0.09 promoter hydrolase, 1 4_P02 PTH2
08XJ GRMZM2G17366 Kinesin, motor DtA-WW PZE-108071309 8 1.23E+08 4.67E-05 0.10 Exon 1 6_P01 region
08XJ AC199790.4_FGT DtS-WS PZE-108084629 8 1.41E+08 6.71E-05 0.12 promoter unknown 1 002
08XJ DtS-WW PZE-108087869 8 1.44E+08 4.24E-05 0.10 NA 1
Protein of GRMZM2G13058 unknown KerN-WW 07HN PZE-108091526 8 1.47E+08 3.92E-05 0.10 intron 6_P01 function DUF1296
Myb-like DNA- 08XJ GRMZM2G06770 binding GY-WS PZE-108100953 8 1.55E+08 9.01E-05 0.09 promoter 1 2_P01 region, SHAQKYF class
GRMZM2G01017 Antifreeze ASI-WW 07XJ PZE-108105473 8 1.59E+08 2.58E-06 0.21 Exon 6_P01 protein, type I
U2 auxiliary GRMZM2G10749 DtA-WS 08HN SYN1030 8 1.61E+08 6.26E-05 0.09 intron factor small 1_P01 subunit Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
U2 auxiliary GRMZM2G10749 DtS-WS 08HN SYN1030 8 1.61E+08 3.25E-05 0.10 intron factor small 1_P01 subunit
GRMZM2G10749 DtA-WS 08HN PZE-108108866 8 1.61E+08 9.52E-06 0.11 intron unknown 8_P01
GRMZM2G10749 DtS-WS 08HN PZE-108108866 8 1.61E+08 4.33E-06 0.13 intron unknown 8_P01
GRMZM2G03135 Armadillo- DtA-WS 08HN SYN1000 8 1.61E+08 5.35E-05 0.09 5 UTR 2_P02 type fold
GRMZM2G56916 DtA-WS 08HN PZE-108117143 8 1.65E+08 1.64E-05 0.12 promoter unknown 4_P01
GRMZM2G56916 DtS-WS 08HN PZE-108117143 8 1.65E+08 7.68E-05 0.11 promoter unknown 4_P01
PlaH-WS 06HN PZE-108120295 8 1.67E+08 2.00E-05 0.13 NA
GRMZM2G04641 PlaH-WS 06HN SYN21962 8 1.68E+08 3.75E-05 0.13 intron unknown 8_P01
Low density lipoprotein- GRMZM2G14903 DtS-WS 08HN PZE-108134760 8 1.73E+08 9.36E-05 0.09 intron receptor, class 1_P01 A, cysteine- rich Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Leucine-rich repeat, 08XJ GRMZM2G11028 DtA-WW SYN24557 9 2.33E+06 4.00E-05 0.10 Exon ribonuclease 1 9_P01 inhibitor subtype
GRMZM2G16000 Transcriptiona ASI-WW 07XJ PZE-109009987 9 1.10E+07 9.75E-05 0.09 Exon 5_P01 l factor B3
08XJ GRMZM2G00181 Ribosomal ASI-WW PZE-109045108 9 7.35E+07 5.29E-05 0.12 intron 1 6_P01 protein L5
Phospholipase C, 08XJ GRMZM2G15776 ASI-WW PZE-109080182 9 1.24E+08 2.38E-05 0.12 intron phosphoinosit 1 0_P01 ol-specific, C- terminal (PLC)
GRMZM2G47335 KerN-WW 07XJ PZE-109086365 9 1.31E+08 8.25E-05 0.11 5 UTR Prion protein 6_P01
GRMZM2G47924 Tyrosine DtS-WS 08HN PZE-109094249 9 1.36E+08 9.13E-05 0.09 Exon 3_P01 protein kinase
08XJ GRMZM2G34899 DtS-WW PZE-110015128 10 1.48E+07 2.52E-05 0.10 promoter WD40 repeat 1 2_P01
08XJ DtS-WW PZE-110015490 10 1.53E+07 9.40E-05 0.11 NA 1 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
08XJ DtS-WW PZE-110017862 10 2.03E+07 8.98E-05 0.09 NA 1
08XJ DtS-WS PZE-110017862 10 2.03E+07 9.95E-05 0.09 NA 1
Pyruvate phosphate 08XJ GRMZM2G04096 DtS-WW PZE-110017902 10 2.04E+07 8.98E-05 0.09 intron dikinase, 1 8_P03 PEP/pyruvate- binding
Pyruvate phosphate 08XJ GRMZM2G04096 DtS-WS PZE-110017902 10 2.04E+07 9.95E-05 0.09 intron dikinase, 1 8_P03 PEP/pyruvate- binding
Pyruvate phosphate 08XJ GRMZM2G04096 DtS-WW SYN26509 10 2.04E+07 8.13E-05 0.09 intron dikinase, 1 8_P03 PEP/pyruvate- binding
Pyruvate phosphate 08XJ GRMZM2G04096 DtS-WS SYN26509 10 2.04E+07 8.59E-05 0.09 intron dikinase, 1 8_P03 PEP/pyruvate- binding Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Pyruvate phosphate 08XJ GRMZM2G04096 DtS-WS SYN26514 10 2.05E+07 9.95E-05 0.09 Exon dikinase, 1 8_P02 PEP/pyruvate- binding
Pyruvate phosphate 08XJ GRMZM2G04096 DtS-WW SYN26514 10 2.05E+07 8.98E-05 0.09 Exon dikinase, 1 8_P02 PEP/pyruvate- binding
Aldehyde 08XJ GRMZM2G33136 dehydrogenas DtS-WW PZE-110017983 10 2.08E+07 3.50E-05 0.10 intron 1 8_P01 e, conserved site
Aldehyde 08XJ GRMZM2G33136 dehydrogenas DtS-WS PZE-110017983 10 2.08E+07 3.41E-05 0.10 intron 1 8_P01 e, conserved site
GRMZM2G40437 Armadillo- KerN-WW 07XJ SYN17646 10 2.22E+07 8.85E-05 0.09 Exon 7_P01 type fold
EarN-WW 07HN PZE-110036801 10 7.01E+07 7.18E-05 0.09 NA
EarN-WW 07HN PZE-110036804 10 7.01E+07 7.18E-05 0.09 NA
EarN-WW 07HN PZE-110036805 10 7.01E+07 7.18E-05 0.09 NA Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
GRMZM2G33714 GY-WS 09XJ PZE-110047457 10 8.85E+07 5.00E-05 0.10 promoter unknown 3_P01
Peptidase S1C, 08XJ GRMZM2G06359 DtS-WS PZE-110047994 10 8.99E+07 2.79E-05 0.12 3 UTR HrtA/DegP2/Q 1 2_P01 /S
08XJ DtS-WW PZE-110049040 10 9.18E+07 9.19E-05 0.10 NA 1
ASI-WW 07XJ PZE-110049494 10 9.30E+07 1.93E-06 0.19 NA
08XJ ASI-WW PZE-110049494 10 9.30E+07 4.87E-06 0.20 NA 1
08XJ DtS-WW PZE-110049494 10 9.30E+07 5.79E-05 0.17 NA 1
08XJ GRMZM2G14268 TonB box, ASI-WW PZE-110050293 10 9.48E+07 2.65E-06 0.17 Exon 1 0_P01 conserved site
Protein of 08XJ GRMZM2G17171 unknown ASI-WW PZE-110050326 10 9.49E+07 2.17E-05 0.15 intron 1 6_P01 function DUF1295
Protein of 08XJ GRMZM2G17171 unknown DtA-WW PZE-110050326 10 9.49E+07 4.39E-05 0.13 intron 1 6_P01 function DUF1295 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Protein of GRMZM2G17171 unknown DtS-WW 07XJ PZE-110050326 10 9.49E+07 5.76E-06 0.15 intron 6_P01 function DUF1295
Protein of 08XJ GRMZM2G17171 unknown DtS-WW PZE-110050326 10 9.49E+07 4.76E-08 0.22 intron 1 6_P01 function DUF1295
Protein of GRMZM2G17171 unknown DtA-WS 07XJ PZE-110050326 10 9.49E+07 1.72E-05 0.14 intron 6_P01 function DUF1295
Protein of GRMZM2G17171 unknown DtS-WS 07XJ PZE-110050326 10 9.49E+07 1.38E-06 0.18 intron 6_P01 function DUF1295
Protein of 08XJ GRMZM2G17171 unknown DtS-WS PZE-110050326 10 9.49E+07 3.04E-06 0.16 intron 1 6_P01 function DUF1295
Peptidase S24, GRMZM2G00071 DtA-WW 07XJ PZE-110050720 10 9.55E+07 6.51E-05 0.09 intron S26A, S26B 0_P01 and S26C Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
DtS-WS 07XJ PZE-110051078 10 9.62E+07 4.12E-05 0.10 NA
Mitochondrial GRMZM2G04254 brown fat DtS-WS 07XJ PZE-110051090 10 9.62E+07 4.55E-05 0.10 5 UTR 1_P01 uncoupling protein
GRMZM2G39778 DtS-WW 07XJ PZE-110051604 10 9.72E+07 3.52E-05 0.12 Exon unknown 8_P01
08XJ GRMZM2G39778 DtS-WW PZE-110051604 10 9.72E+07 2.09E-07 0.18 Exon unknown 1 8_P01
GRMZM2G39778 DtA-WS 07XJ PZE-110051604 10 9.72E+07 7.82E-05 0.11 Exon unknown 8_P01
GRMZM2G39778 DtS-WS 07XJ PZE-110051604 10 9.72E+07 5.84E-06 0.14 Exon unknown 8_P01
08XJ GRMZM2G39778 DtS-WS PZE-110051604 10 9.72E+07 1.03E-05 0.14 Exon unknown 1 8_P01
08XJ GRMZM2G39778 ASI-WW PZE-110051604 10 9.72E+07 1.05E-06 0.17 Exon unknown 1 8_P01
08XJ GRMZM2G09917 DtS-WW PZE-110051617 10 9.72E+07 4.61E-06 0.14 Exon unknown 1 2_P01
08XJ GRMZM2G09917 ASI-WW PZE-110051621 10 9.72E+07 9.42E-07 0.17 Exon unknown 1 2_P01 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
GRMZM2G09917 DtS-WW 07XJ PZE-110051621 10 9.72E+07 1.73E-05 0.12 Exon unknown 2_P01
08XJ GRMZM2G09917 DtS-WW PZE-110051621 10 9.72E+07 2.24E-07 0.18 Exon unknown 1 2_P01
GRMZM2G09917 DtS-WS 07XJ PZE-110051621 10 9.72E+07 6.30E-06 0.14 Exon unknown 2_P01
08XJ GRMZM2G09917 DtS-WS PZE-110051621 10 9.72E+07 4.75E-06 0.15 Exon unknown 1 2_P01
Proteinase 08XJ GRMZM2G07531 DtS-WW PZE-110051945 10 9.79E+07 5.43E-05 0.12 promoter inhibitor I12, 1 5_P01 Bowman-Birk
Proteinase GRMZM2G07531 DtS-WS 07XJ PZE-110051945 10 9.79E+07 2.14E-05 0.13 promoter inhibitor I12, 5_P01 Bowman-Birk
08XJ DtS-WW PZE-110053302 10 1.00E+08 1.13E-05 0.11 NA 1
08XJ ASI-WW PZE-110054846 10 1.05E+08 4.75E-05 0.10 NA 1
08XJ DtS-WW PZE-110055733 10 1.07E+08 9.46E-05 0.09 NA 1
08XJ ASI-WW PZE-110060926 10 1.15E+08 5.50E-05 0.12 NA 1 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
DtS-WW 07XJ PZE-110060926 10 1.15E+08 2.23E-05 0.12 NA
08XJ DtS-WW PZE-110060926 10 1.15E+08 1.65E-07 0.19 NA 1
08XJ DtS-WS PZE-110060926 10 1.15E+08 3.40E-05 0.12 NA 1
ASI-WW 07XJ PZE-110061159 10 1.15E+08 2.17E-05 0.14 NA
GY-WW 07HN PZE-110063916 10 1.19E+08 6.17E-05 0.09 NA
Uncharacteris ed protein family 08XJ GRMZM2G08057 UPF0497, ASI-WW PZE-110066634 10 1.23E+08 4.25E-06 0.15 promoter 1 5_P01 trans- membrane plant subgroup
Uncharacteris ed protein family 08XJ GRMZM2G08057 UPF0497, DtS-WW PZE-110066634 10 1.23E+08 8.01E-05 0.11 promoter 1 5_P01 trans- membrane plant subgroup Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
08XJ PUT-163a- GRMZM2G10711 DtS-WW 10 1.23E+08 2.52E-05 0.10 Exon Prohibitin 1 148967626-627 4_P01
DtA-WS 07XJ PZE-110069086 10 1.26E+08 3.86E-05 0.10 NA
S- adenosylmethi 08XJ GRMZM2G12563 ASI-WW PZE-110071651 10 1.28E+08 2.58E-05 0.11 5 UTR onine 1 5_P01 decarboxylase subgroup
DEAD-like GRMZM2G01831 KerN-WW 07XJ SYN4817 10 1.34E+08 5.66E-05 0.11 Exon helicase, N- 4_P01 terminal
08XJ GRMZM2G10419 PlaH-WW PZE-110103147 10 1.46E+08 8.05E-05 0.10 5 UTR unknown 1 9_P01 Supplementary Table 3 SNPs associated with different traits or in different environment under WS condition Env Trait SNP Chr Site P R2 Gene Name Gene Feature Function .
CBL- interacting 08X GRMZM2G31576 DtA-WS PZE-103126852 3 1.82E+08 7.88E-06 0.12 intron serine/threoni J1 9_P01 ne-protein kinase 15
CBL- interacting 07X GRMZM2G31576 DtA-WS PZE-103126852 3 1.82E+08 6.79E-05 0.09 intron serine/threoni J 9_P01 ne-protein kinase 15
CBL- interacting 08X GRMZM2G31576 DtA-WS PZE-103126857 3 1.82E+08 7.88E-06 0.12 5 UTR serine/threoni J1 9_P01 ne-protein kinase 15
CBL- interacting 07X GRMZM2G31576 DtA-WS PZE-103126857 3 1.82E+08 6.79E-05 0.09 5 UTR serine/threoni J 9_P01 ne-protein kinase 15 Env Trait SNP Chr Site P R2 Gene Name Gene Feature Function .
Alpha-1,4- 08X PUT-163a- GRMZM2G08732 glucan-protein PlaH-WS 5 1.87E+07 3.74E-05 0.12 Exon J1 16925922-1125 6_P01 synthase, UDP-forming
Alpha-1,4- 07X PUT-163a- GRMZM2G08732 glucan-protein PlaH-WS 5 1.87E+07 8.70E-05 0.11 Exon J 16925922-1125 6_P01 synthase, UDP-forming
08 GRMZM2G10749 DtA-WS PZE-108108866 8 1.61E+08 9.52E-06 0.11 intron unknown HN 8_P01
08 GRMZM2G10749 DtS-WS PZE-108108866 8 1.61E+08 4.33E-06 0.13 intron unknown HN 8_P01
zinc finger CCCH type 08 GRMZM2G10749 domain- DtA-WS SYN1030 8 1.61E+08 6.26E-05 0.09 intron HN 1_P01 containing protein ZFN- like 2 Env Trait SNP Chr Site P R2 Gene Name Gene Feature Function .
zinc finger CCCH type 08 GRMZM2G10749 domain- DtS-WS SYN1030 8 1.61E+08 3.25E-05 0.1 intron HN 1_P01 containing protein ZFN- like 3
Protein of 08 GRMZM2G17171 unknown DtA-WS PZE-108117143 8 1.65E+08 1.64E-05 0.12 intron HN 6_P01 function DUF1295
Protein of 08 GRMZM2G17171 unknown DtS-WS PZE-108117143 8 1.65E+08 7.68E-05 0.11 intron HN 6_P01 function DUF1295
Protein of 07X GRMZM2G17171 unknown DtA-WS PZE-110050326 10 9.49E+07 1.72E-05 0.14 intron J 6_P01 function DUF1295
Protein of 07X GRMZM2G17171 unknown DtS-WS PZE-110050326 10 9.49E+07 1.38E-06 0.18 intron J 6_P01 function DUF1295 Env Trait SNP Chr Site P R2 Gene Name Gene Feature Function .
Protein of 08X GRMZM2G17171 unknown DtS-WS PZE-110050326 10 9.49E+07 3.04E-06 0.16 intron J1 6_P01 function DUF1295
07X GRMZM2G39778 DtA-WS PZE-110051604 10 9.72E+07 7.82E-05 0.11 Exon unknown J 8_P01
07X GRMZM2G39778 DtS-WS PZE-110051604 10 9.72E+07 5.84E-06 0.14 Exon unknown J 8_P01
08X GRMZM2G39778 DtS-WS PZE-110051604 10 9.72E+07 1.03E-05 0.14 Exon unknown J1 8_P01
Protein of 07X GRMZM2G17171 unknown DtS-WS PZE-110050326 10 9.49E+07 1.38E-06 0.18 intron J 6_P01 function DUF1295
Protein of 08X GRMZM2G17171 unknown DtS-WS PZE-110050326 10 9.49E+07 3.04E-06 0.16 intron J1 6_P01 function DUF1295
07X GRMZM2G39778 DtS-WS PZE-110051604 10 9.72E+07 5.84E-06 0.14 Exon unknown J 8_P01
08X GRMZM2G39778 DtS-WS PZE-110051604 10 9.72E+07 1.03E-05 0.14 Exon unknown J1 8_P01 Env Trait SNP Chr Site P R2 Gene Name Gene Feature Function .
07X GRMZM2G09917 DtS-WS PZE-110051621 10 9.72E+07 6.30E-06 0.14 Exon unknown J 2_P01
08X GRMZM2G09917 DtS-WS PZE-110051621 10 9.72E+07 4.75E-06 0.15 Exon unknown J1 2_P01
Supplementary Table 4 SNPs detected under both water conditions Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
CBL- interacting 08XJ GRMZM2G315769_P DtA-WW PZE-103126852 3 1.82E+08 2.90E-05 0.10 intron serine/threoni 1 01 ne-protein kinase 15
CBL- interacting GRMZM2G315769_P DtA-WS 07XJ PZE-103126852 3 1.82E+08 6.79E-05 0.09 intron serine/threoni 01 ne-protein kinase 15
CBL- interacting 08XJ GRMZM2G315769_P DtA-WS PZE-103126852 3 1.82E+08 7.88E-06 0.12 intron serine/threoni 1 01 ne-protein kinase 15 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
CBL- interacting 08XJ GRMZM2G315769_P DtA-WW PZE-103126857 3 1.82E+08 2.90E-05 0.10 5 UTR serine/threoni 1 01 ne-protein kinase 15
CBL- interacting GRMZM2G315769_P DtA-WS 07XJ PZE-103126857 3 1.82E+08 6.79E-05 0.09 5 UTR serine/threoni 01 ne-protein kinase 15
CBL- interacting 08XJ GRMZM2G315769_P DtA-WS PZE-103126857 3 1.82E+08 7.88E-06 0.12 5 UTR serine/threoni 1 01 ne-protein kinase 15
Alpha-1,4- PUT-163a- GRMZM2G087326_P glucan-protein PlaH-WS 07XJ 5 1.87E+07 8.70E-05 0.11 Exon 16925922-1125 01 synthase, UDP-forming
Alpha-1,4- 08XJ PUT-163a- GRMZM2G087326_P glucan-protein PlaH-WS 5 1.87E+07 3.74E-05 0.12 Exon 1 16925922-1125 01 synthase, UDP-forming Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Alpha-1,4- 08XJ PUT-163a- GRMZM2G087326_P glucan-protein PlaH-WW 5 1.87E+07 3.61E-05 0.12 Exon 1 16925922-1125 01 synthase, UDP-forming
GRMZM2G342243_P Harpin- DtA-WW 09XJ SYN11613 6 7.77E+07 1.36E-05 0.12 Exon 01 induced 1
08XJ GRMZM2G342243_P Harpin- DtS-WS SYN11613 6 7.77E+07 6.71E-05 0.09 Exon 1 01 induced 1
Peptidyl-tRNA 08XJ GRMZM2G127844_P DtA-WW PZE-108071041 8 1.23E+08 2.47E-05 0.10 promoter hydrolase, 1 02 PTH2
Peptidyl-tRNA 08XJ GRMZM2G127844_P PlaH-WS PZE-108071041 8 1.23E+08 7.63E-05 0.09 promoter hydrolase, 1 02 PTH2
08XJ DtS-WW PZE-110017862 10 2.03E+07 8.98E-05 0.09 NA 1
08XJ DtS-WS PZE-110017862 10 2.03E+07 9.95E-05 0.09 NA 1
Pyruvate phosphate 08XJ GRMZM2G040968_P DtS-WW PZE-110017902 10 2.04E+07 8.98E-05 0.09 intron dikinase, 1 03 PEP/pyruvate- binding Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Pyruvate phosphate 08XJ GRMZM2G040968_P DtS-WS PZE-110017902 10 2.04E+07 9.95E-05 0.09 intron dikinase, 1 03 PEP/pyruvate- binding
Aldehyde 08XJ GRMZM2G331368_P dehydrogenas DtS-WW PZE-110017983 10 2.08E+07 3.50E-05 0.10 intron 1 01 e, conserved site
Aldehyde 08XJ GRMZM2G331368_P dehydrogenas DtS-WS PZE-110017983 10 2.08E+07 3.41E-05 0.10 intron 1 01 e, conserved site
Protein of 08XJ GRMZM2G171716_P unknown ASI-WW PZE-110050326 10 9.49E+07 2.17E-05 0.15 intron 1 01 function DUF1295
Protein of 08XJ GRMZM2G171716_P unknown DtA-WW PZE-110050326 10 9.49E+07 4.39E-05 0.13 intron 1 01 function DUF1295 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Protein of GRMZM2G171716_P unknown DtS-WW 07XJ PZE-110050326 10 9.49E+07 5.76E-06 0.15 intron 01 function DUF1295
Protein of 08XJ GRMZM2G171716_P unknown DtS-WW PZE-110050326 10 9.49E+07 4.76E-08 0.22 intron 1 01 function DUF1295
Protein of GRMZM2G171716_P unknown DtA-WS 07XJ PZE-110050326 10 9.49E+07 1.72E-05 0.14 intron 01 function DUF1295
Protein of GRMZM2G171716_P unknown DtS-WS 07XJ PZE-110050326 10 9.49E+07 1.38E-06 0.18 intron 01 function DUF1295
Protein of 08XJ GRMZM2G171716_P unknown DtS-WS PZE-110050326 10 9.49E+07 3.04E-06 0.16 intron 1 01 function DUF1295
08XJ GRMZM2G397788_P ASI-WW PZE-110051604 10 9.72E+07 1.05E-06 0.17 Exon unknown 1 01 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
GRMZM2G397788_P DtS-WW 07XJ PZE-110051604 10 9.72E+07 3.52E-05 0.12 Exon unknown 01
08XJ GRMZM2G397788_P DtS-WW PZE-110051604 10 9.72E+07 2.09E-07 0.18 Exon unknown 1 01
GRMZM2G397788_P DtA-WS 07XJ PZE-110051604 10 9.72E+07 7.82E-05 0.11 Exon unknown 01
GRMZM2G397788_P DtS-WS 07XJ PZE-110051604 10 9.72E+07 5.84E-06 0.14 Exon unknown 01
08XJ GRMZM2G397788_P DtS-WS PZE-110051604 10 9.72E+07 1.03E-05 0.14 Exon unknown 1 01
08XJ GRMZM2G099172_P ASI-WW PZE-110051621 10 9.72E+07 9.42E-07 0.17 Exon unknown 1 01
GRMZM2G099172_P DtS-WW 07XJ PZE-110051621 10 9.72E+07 1.73E-05 0.12 Exon unknown 01
08XJ GRMZM2G099172_P DtS-WW PZE-110051621 10 9.72E+07 2.24E-07 0.18 Exon unknown 1 01
GRMZM2G099172_P DtS-WS 07XJ PZE-110051621 10 9.72E+07 6.30E-06 0.14 Exon unknown 01
08XJ GRMZM2G099172_P DtS-WS PZE-110051621 10 9.72E+07 4.75E-06 0.15 Exon unknown 1 01 Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Proteinase 08XJ GRMZM2G075315_P DtS-WW PZE-110051945 10 9.79E+07 5.43E-05 0.12 promoter inhibitor I12, 1 01 Bowman-Birk
Proteinase GRMZM2G075315_P DtS-WS 07XJ PZE-110051945 10 9.79E+07 2.14E-05 0.13 promoter inhibitor I12, 01 Bowman-Birk
08XJ ASI-WW PZE-110060926 10 1.15E+08 5.50E-05 0.12 NA 1
DtS-WW 07XJ PZE-110060926 10 1.15E+08 2.23E-05 0.12 NA
08XJ DtS-WW PZE-110060926 10 1.15E+08 1.65E-07 0.19 NA 1
08XJ DtS-WS PZE-110060926 10 1.15E+08 3.40E-05 0.12 NA 1
Pyruvate phosphate 08XJ GRMZM2G040968_P DtS-WW SYN26509 10 2.04E+07 8.13E-05 0.09 intron dikinase, 1 03 PEP/pyruvate- binding
Pyruvate phosphate 08XJ GRMZM2G040968_P DtS-WS SYN26509 10 2.04E+07 8.59E-05 0.09 intron dikinase, 1 03 PEP/pyruvate- binding Trait Env. SNP Chr Site P R2 Gene Name Gene Feature Function
Pyruvate phosphate 08XJ GRMZM2G040968_P DtS-WS SYN26514 10 2.05E+07 9.95E-05 0.09 Exon dikinase, 1 02 PEP/pyruvate- binding
Pyruvate phosphate 08XJ GRMZM2G040968_P DtS-WW SYN26514 10 2.05E+07 8.98E-05 0.09 Exon dikinase, 1 02 PEP/pyruvate- binding Supplementary Fig.1 Distribution frequencies of seven traits among maize inbred lines under both WS and WW conditions. The phenotypic traits are abbreviated as EarN (total ears number per plot), KerN (kernel number per row), PlaH (plant height), ASI (anthesis-silking interval), DtA (days to anthesis), DtS (days to silking), and GY (grain yield). Treatments are abbreviated as WW (well-watered regime) and WS (water-stressed regime).