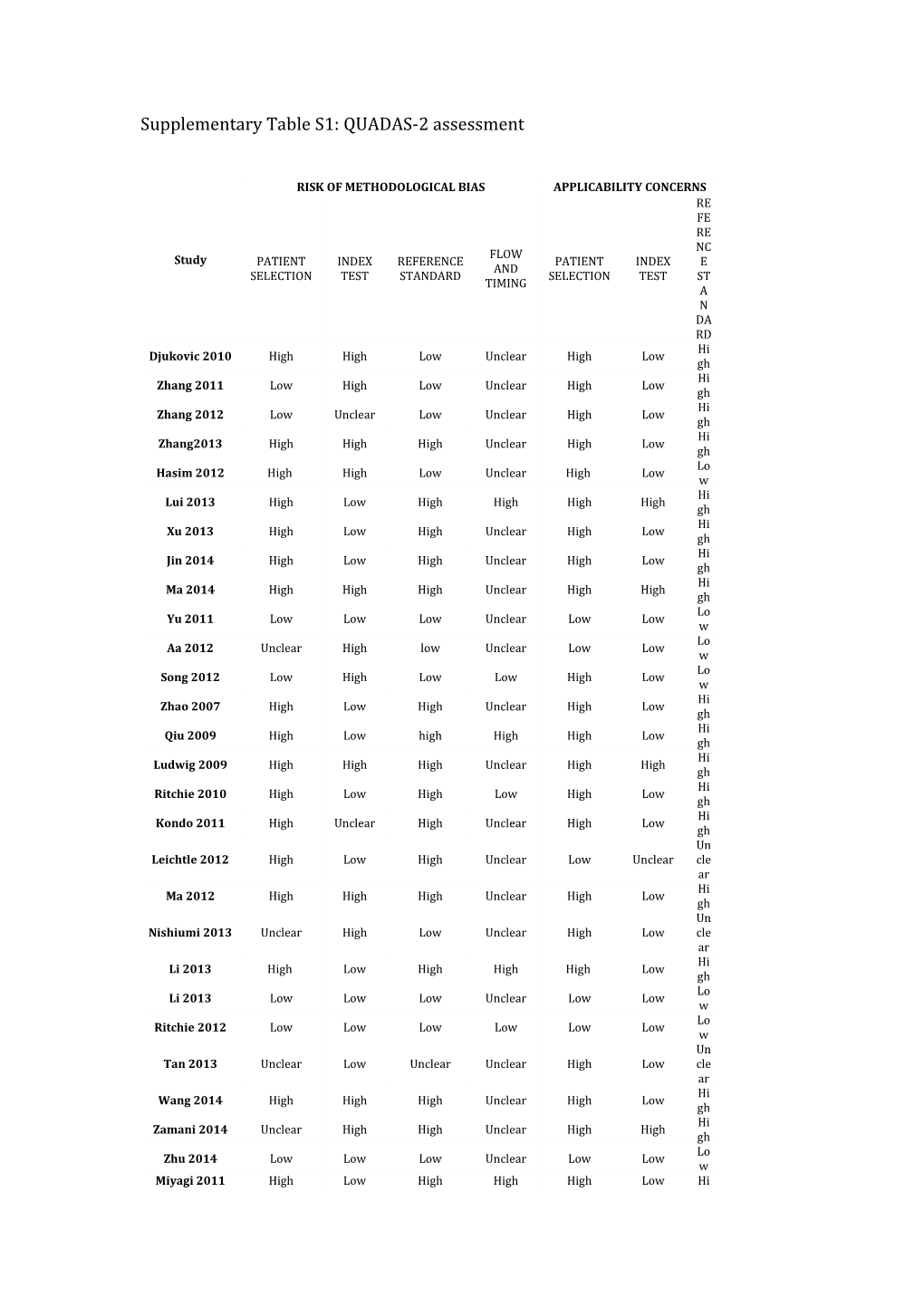
Supplementary Table S1: QUADAS-2 assessment
RISK OF METHODOLOGICAL BIAS APPLICABILITY CONCERNS RE FE RE NC FLOW Study PATIENT INDEX REFERENCE PATIENT INDEX E AND SELECTION TEST STANDARD SELECTION TEST ST TIMING A N DA RD Hi Djukovic 2010 High High Low Unclear High Low gh Hi Zhang 2011 Low High Low Unclear High Low gh Hi Zhang 2012 Low Unclear Low Unclear High Low gh Hi Zhang2013 High High High Unclear High Low gh Lo Hasim 2012 High High Low Unclear High Low w Hi Lui 2013 High Low High High High High gh Hi Xu 2013 High Low High Unclear High Low gh Hi Jin 2014 High Low High Unclear High Low gh Hi Ma 2014 High High High Unclear High High gh Lo Yu 2011 Low Low Low Unclear Low Low w Lo Aa 2012 Unclear High low Unclear Low Low w Lo Song 2012 Low High Low Low High Low w Hi Zhao 2007 High Low High Unclear High Low gh Hi Qiu 2009 High Low high High High Low gh Hi Ludwig 2009 High High High Unclear High High gh Hi Ritchie 2010 High Low High Low High Low gh Hi Kondo 2011 High Unclear High Unclear High Low gh Un Leichtle 2012 High Low High Unclear Low Unclear cle ar Hi Ma 2012 High High High Unclear High Low gh Un Nishiumi 2013 Unclear High Low Unclear High Low cle ar Hi Li 2013 High Low High High High Low gh Lo Li 2013 Low Low Low Unclear Low Low w Lo Ritchie 2012 Low Low Low Low Low Low w Un Tan 2013 Unclear Low Unclear Unclear High Low cle ar Hi Wang 2014 High High High Unclear High Low gh Hi Zamani 2014 Unclear High High Unclear High High gh Lo Zhu 2014 Low Low Low Unclear Low Low w Miyagi 2011 High Low High High High Low Hi gh Lo Ikeda 2012 High High Low Unclear High Low w Supplementary Table S2: Index test methodological quality
Was compound identification Were standards method more Were there any Potential for used for than electronic quality control methodological quantification reference library measures? bias (MS only)? (e.g. isotope standards, TOF etc) (MS only)?
Djukovic 2010 Yes Yes No High Zhang 2011 - - No High Zhang 2012 Yes Yes Yes Low Zhang2013 No No No High Hasim 2012 - - No High Lui 2013 Yes Yes Yes Low Xu 2013 Yes Yes Yes Low Jin 2014 Yes Yes Yes Low Ma 2014 No Yes Yes High Yu 2011 Yes Yes Yes Low Aa 2012 Yes Yes No High Song 2012 Yes No Yes High Zhao 2007 Yes Yes Yes Low Qiu 2009 Yes Yes Yes Low Ludwig 2009 - - No High Ritchie 2010 Yes Yes Yes Low Kondo 2011 Yes Yes Yes Low Leichtle 2012 Yes Yes Yes Low Ma 2012 No Yes Yes High Nishiumi 2013 No Yes Yes High Li 2013 Yes Yes Yes Low Li 2013 Yes Yes Yes Low Ritchie 2013 Yes Yes Yes Low Tan 2013 Yes Yes Yes Low Wang 2014 No No No High Zamani 2014 - - No High Zhu 2014 Yes Yes Yes Low Miyagi 2011 Yes Yes Yes Low Ikeda 2012 No Yes Yes High Supplementary Table S3: STARD checklist assessment
Did the study:
Identify the a rticle as a s tudy of dia gnosti c 0 0 0 1 0 0 0 1 0 0 0 1 0 0 1 0 1 0 0 1 0 0 0 0 1 1 0 1 0 a ccura cy?
Sta te the res earch ai ms ? 0 0 0 1 1 0 1 1 0 0 1 1 0 0 0 0 1 0 0 1 0 0 0 0 1 1 0 1 0
Des cribe popul ation s el ection criteria ? 1 1 1 1 1 0 1 1 1 1 0 0 1 1 1 1 1 1 1 1 1 1 1 0 1 0 1 1 1
Describe pa ti ent s el ection criteria? 1 1 1 1 1 0 1 1 1 1 1 1 0 1 1 1 1 1 1 1 1 0 1 0 1 1 0 1 1
Sta te pa ttern of recruitment (e.g. cons ecutive)? 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1 0 0 0 0 0 0 0 0 0 0 1 1
State if da ta before or after testing? 0 1 0 1 1 0 1 1 0 1 0 0 0 1 1 0 1 1 1 0 0 1 1 0 0 1 0 1 1
Sta te rati ona le of reference s tandard? 1 0 1 1 0 0 0 1 0 0 1 1 0 1 1 1 1 1 0 0 1 1 1 0 1 1 0 1 1
Des cribe the technica l deta il s of the index 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 1 1 1 test? Define units a nd/or cut-offs for the i ndex tes t 0 1 1 0 1 0 1 1 1 0 0 0 0 1 1 0 1 1 0 1 1 1 0 0 1 1 1 1 1 and reference s tanda rd?
Des cribe who ca rried out tes ting? 0 0 0 0 0 0 0 0 1 0 0 1 0 1 1 0 1 0 0 0 1 0 0 0 1 1 1 1 0
Des cribe whether or not thos e carrying out 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 1 1 0 1 1 1 1 1 1 1 1 1 1 1 testing were bl inded? Des cribe methods of determining test accuracy 1 0 0 0 0 1 1 0 1 0 1 0 1 1 1 0 1 1 0 0 0 1 0 0 1 1 1 1 1 and s ta ti stica l uncerta inty? Des cribe methods for ca lcula ti ng tes t NA NA NA 0 0 0 1 1 NA NA 0 0 NA 1 1 NA 1 1 NA 1 0 1 NA 0 1 1 1 1 1 reproduci bi lity, if done?
Sta te when the study was performed? 0 0 1 0 0 0 0 1 0 1 0 0 1 1 1 0 0 1 0 0 1 0 1 0 0 0 0 1 1
Des cribed bas i c demographics of pa rtici pants ? 0 0 0 1 1 1 1 1 0 0 0 0 1 1 1 0 1 1 1 1 0 0 1 0 0 1 0 1 1
Sta te the number of el igible pa tients not 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 incl uded, and the reas ons why? Give the time interva l between the index tes t 1 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 and reference s tanda rd? Des cribe the severity of the dis eas e in 0 0 1 0 0 1 1 1 0 0 0 0 1 1 1 1 1 1 1 1 1 1 0 0 0 1 0 1 1 participants wi th defined criteria ? Provide cross -ta bul a tion etc. of patient 0 0 0 0 0 0 1 1 1 0 0 0 0 0 1 0 0 0 0 1 0 0 0 0 0 0 0 1 0 a ss ignment by tes ts ?
Des cribe advers e events during testing? 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Provide estimates of dia gnos tic accuracy a nd 0 0 0 1 0 0 1 0 0 0 0 0 0 1 1 0 1 1 1 0 0 0 0 0 1 0 1 1 0 s ta tis tica l uncerta inty? Des cribe how uncla ssi fied pa tients a nd 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 1 0 outliers were a nalysed? Describe es timates of accuracy between NA NA NA NA NA 0 1 NA 1 NA 0 0 NA 0 1 NA NA NA NA NA 0 NA NA NA 1 1 1 NA NA s ubgroups or centres , if done? Provide es tima tes of tes t reproducibil ity, if NA NA NA NA NA 0 NA 0 0 NA 0 0 NA NA 1 NA NA NA NA 1 0 NA NA NA 1 0 0 NA NA done? Dis cuss the clinica l a pplica bility of the study 1 1 1 1 1 1 1 1 1 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 1 fi ndings ? Supplementary Table S4: All significant metabolites
All listed metabolites by disease type, reported frequency and KEGG Compound assignment number. Where a metabolite is not KEGG-assigned, Human Metabolite DataBase assignments are given.
Freq EAC UP . Ref. EAC DOWN Freq. Ref. 3-hydroxybutyric acid 2 C01089 1-methyladenosine, 1 C02494 aspartic acid 1 C00402 2,2,dimethylguanosine 1 HMDB04824 carnitine 1 C00487 5-hydroxytryptophan 1 C00643 citrate 1 C00158 a-glucose 1 C00267 creatine 2 C00300 acetate 1 C00033 creatinine 1 C00791 cytidine 1 C00475 cysteine 1 C00097 leucine/isoleucine 1 C00123 glucose 1 C00031 linoleic acid 1 C01595 glutamic acid 2 C00217 linolenic acid 1 CO6427 histidine 1 C00135 methionine 2 C00073 lactic acid 2 C00256 myristic acid 1 C06424 leucine/isoleucine 1 C00123 N2-methylguanosine 1 C20674 lysine 2 C00047 tryptophan 2 C00078 phenylalanine 1 C00355 tyrosine 2 C00082 uridine 1 C00299 Valine 1 C00183
Total 15 metabolites Total 15 metabolites ESCC UP Freq. Ref. ESCC DOWN Freq. Ref. 1-monooleoylglycerol 1 n/a 1,5-anhydroglucitol 1 C07326 3-hydroxybutyric acid 2 C01089 2-ketoisocaproic acid 1 C03467 alanine 1 C00993 2-ketoisovaleric acid 1 C06255 aspartic acid 2 C00402 3-methyl-2oxovaleric acid 1 C06255 creatinine 1 C00791 5-B-cyprinol sulphate 1 C05468 hypotaurine 1 C00591 a-tocopherol 1 C02477 l-urobilinogen 1 C05789 alanine 2 C00041 lactic acid 3 C00256 cholesterol 1 C00187 lactose 1 C00243 citric acid 1 C00158 linoleic acid 1 C01595 cysteine 1 C00097 lithocholate 3-o-glucuronide 1 C03990 desmosine/isodesmosine 1 HMDB00572 lithocholyltaurine 1 C03642 erythritol 1 C00503 maltose 1 C00209 fumaric acid 1 C00122 myristic acid 1 C06424 g-aminobutyric acid 1 C00334 oleic acid 1 C00712 g-tocopherol 1 C02477 palmitelaidic acid 1 HMDB12328 glucose 1 C00031 palmitic acid 1 C00249 glutamic acid 2 C00025 (phosphatidic acid)† 1 n/a glycine 1 C00037 (phosphatidyl 1 n/a glycolic acid 1 C00160 ethanolamine)† (phosphatidylcholine)† 1 n/a histidine 1 C00135 (phosphatidylinositol)† 1 n/a hydroxylamine 1 C00192 phosphatidylserine 1 HMDB12352 iminodiacetic acid 1 C19911 (16:0/14:0) ribose 1 C00672 indolelactic acid 1 C02043 sphinganine 1-phosphate 1 C01120 inositol 1 C00137 leucine/isoleucine 1 C00123 Total 24 metabolites lysoPC (14:0) 1 HMDB10379 lysoPC (20:3) 1 HMDB10393 methionine 1 C00073 methylcysteine 1 HMDB02108 myo-inositol-1-phosphate 1 C01177 phenylalanine 1 C00355 phosphoethanolamine 1 C00346 taurine 1 C00245 threonine 2 C00188 tryptophan 1 C00078 uric acid 1 C00366 valine 1 C00183
Total 37 metabolites
GC UP Freq. Ref. GC DOWN Freq. Ref. 3-hydroxybutyric 1 C01089 1,2,4-benzenetricarboxylic acid 1 n/a acid asparagine 1 C00152 2-amino-4-hydroxy-pteridinone 1 C06313 leucine/isoleucine 1 C00123 benzeneacetonitrile 1 C08345 lysine 1 C00047 cholesta-3,5-diene 1 n/a urate 1 C00366 cholesterol 2 C00187 citrulline 1 C00327 histidine 1 C00135 tryptophan 1 C00078 valine 1 C00183
Total 5 metabolites Total 10 metabolites CRC UP Freq. Ref. CRC DOWN Freq. Ref. 2-aminobutanoic acid 1 C02356 1-hexadecanol 1 C00823 2-hydroxybutyric acid 1 C05984 1,5-anhydroglucitol 1 C07326 12a-hydroxy-3-oxocholadienic 2-octenedioic acid 1 HMDB00341 1 C15569 acid 2-oxobutanoic acid 1 C00109 2-hydroxy-3-methylpentanoic acid 1 HMDB00317 3-hydroxybutyric acid 1 C01089 3-hydroxybutyric acid 1 C01089 O-acylcarnitine 1 C02301 3,4,5-trimethoxycinnamic acid 1 HMDB02511 adenine 1 C00147 4-hydroxyproline 2 C01157 alanine 1 C00041 4-hydroxystyrene 1 C05627 carnitine (18:0) 1 C00318 5-dehydroquinic acid 1 C00944 CPA(18:0/0:0) 1 HMDB07004 5-hydroxytryptamine 1 C00780 creatinine 1 C00791 6-phosphogluconic acid 1 C00345 elaidic acid 1 C01712 aconitate 2 C00417 glyceraldehyde 1 C00577 alanine 3 C00041 glycerol 1 C06037 allantoic acid 1 C00499 glycerol phosphate 1 C00093 arabinose 1 C00259 glycochenodeoxycholic 1 C05466 arabitol 1 C00532 acid hippuric acid 1 C01586 arginine 1 C00062 lactic acid 2 C00256 asparagine 1 C00152 leucine/isoleucine 3 C00123 aspartic acid 3 C00402 linoleic acid 1 C01595 aspartylserine 1 HMDB28762 lysine 2 C00047 benzaldehyde 1 C00261 butyltetramethyl-3,5-decadien-7- lysoPC (18:2) 1 HMDB10386 1 n/a yne lysoPC (20:4) 1 HMDB10395 chenodeoxycholic acid 1 C02528 lysoPC (22:6) 1 HMDB10404 cholic acid 1 C00695 oleamide 1 C19670 citric acid 1 C00158 oleic acid 1 C00712 citrulline 3 C00327 Pyruvic acid 1 C00022 cystamine 1 C01678 serine 1 C00065 cysteine 1 C00097 sphinganine 1 C00836 decanoyl carnitine 1 C03299 tetramethydimethylene- 1 n/a deoxyuridine 1 C00526 cyclohexane xanthosine 1 C01762 dihydroxyacetone 1 C00184 dopamine 1 C03758 Total 31 metabolites eicosatrienoic acid 1 CE2516 erythrotetrofuranose 1 n/a ethylhexanol 1 C02498 fumarate 1 C00122 galactosamine 1 C02262 glucosamine 1 C00329 glucose 1 C00031 glucuronic acid 1 C00191 glutamic acid 3 C00025 glyceric acid 2 C00258 glycine 2 C00037 heptadecanoic acid 1 HMDB02259 hexadecanedioc acid 1 C19615 histidine 5 C00135 hydroquinone 1 C00530 indoleacrylic 1 HMDB00734 acid indoxyl 2 C05658 Inositol 1 C00137 kynurenine 1 C00328 lactic acid 1 C00256 lauric acid 1 C02679 leucine/isoleuci 3 C00123 ne linoleic acid 1 C01595 LPA (18:0/0:0) 1 C00416 lysine 1 C00047 lyso-PAF 1 C04598 lysoPC (14:0) 2 HMDB10379 lysoPC (16:0) 1 HMDB10382 lysoPC 1 HMDB10383 (16:1(9Z)) lysoPC (18:0) 1 HMDB11128 lysoPC (18:1) 1 HMDB10385 lysoPC (18:2) 1 HMDB10386 lysoPC (20:0) 2 HMDB10390 lysoPC (20:4) 1 HMDB10395 lysoPC (22:0) saturated 1 HMDB10398 lysoPC lysoPC (22:6) 1 HMDB10404 lysoPC (P- 1 HMDB10408 18:1(9Z)) malic acid 1 C00149 mannose 1 C00159 Many species of ultralong (C28- 36) polyunsaturated hydroxylated 1 n/a fatty acids deregulated in cancer, most prominently GTA-440 meso-erythritol 1 C00503 methionine 3 C00073 myristic acid 1 C06424 octadecanoic 1 C01530 acid oleamide 2 C19670 ornithine 3 C00515 oxalic acid 1 C00209 p- hydroxybenzoic 1 C00156 acid palmitic acid 1 C00249 palmitic amide 1 HMDB12273 phenol 1 C00146 phenyl methylcarbamat 1 n/a e phenylalanine 2 C00079 putrescine 1 C00134 pyroglutamic 1 C01879 acid pyruvate/oxalac 1 C00022 etate ribitol 1 C00474 ribose 1 C00672 s-benzyl-L- 1 n/a Cysteine sarcosine 1 C00213 serine 2 C00065 Sorbopyranose 1 C08356 sphingosylphos 1 C03640 phorylcholine taurine 1 C00245 threitol 1 C16884 threo-3- hydroxyaspartic 1 C11511 acid threonic acid 1 C01620 threonine 2 C00188 trihydroxycopro 1 HMDB02163 stanoic acid trimethylamine 1 C01104 N-oxide tryptophan 4 C00078 tyrosine 4 C00082 ubiquinone 1 C00399 urea 1 C00086 uridine 1 C00299 valine 4 C00183 xylitol 1 C00379
Total 109 metabolites
Note: †ESCC phospholipids given in brackets as authors did not specify species. lysoPC, lysophosphocholine; CPA, cyclic phosphatidic acid; LPA, lysophophatidic acid.