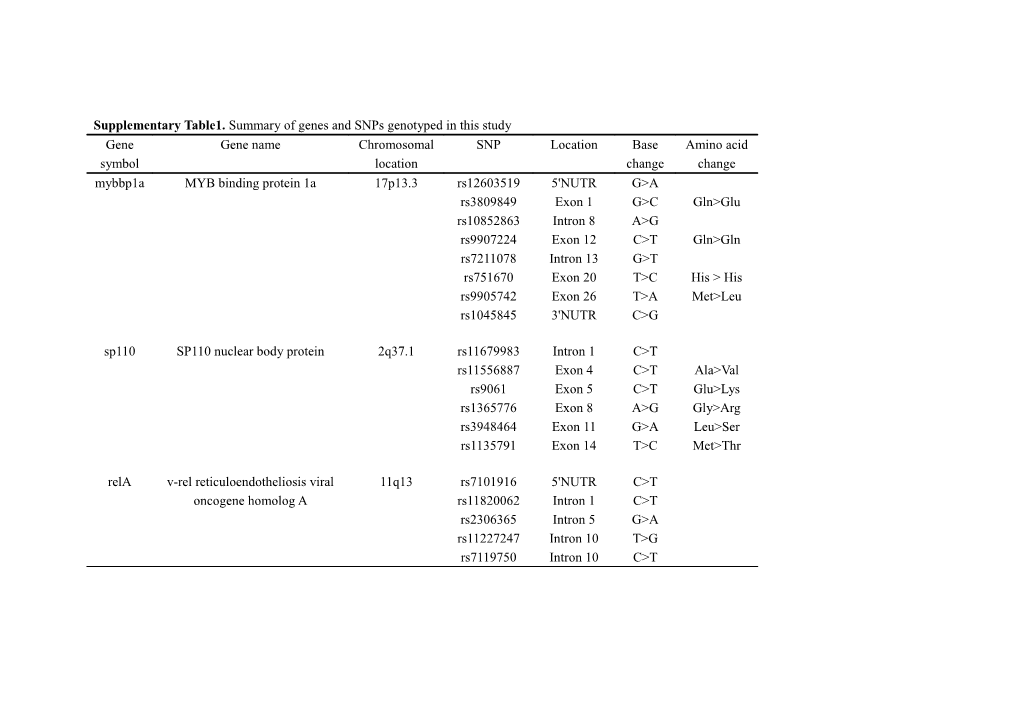
Supplementary Table1. Summary of genes and SNPs genotyped in this study Gene Gene name Chromosomal SNP Location Base Amino acid symbol location change change mybbp1a MYB binding protein 1a 17p13.3 rs12603519 5'NUTR G>A rs3809849 Exon 1 G>C Gln>Glu rs10852863 Intron 8 A>G rs9907224 Exon 12 C>T Gln>Gln rs7211078 Intron 13 G>T rs751670 Exon 20 T>C His > His rs9905742 Exon 26 T>A Met>Leu rs1045845 3'NUTR C>G
sp110 SP110 nuclear body protein 2q37.1 rs11679983 Intron 1 C>T rs11556887 Exon 4 C>T Ala>Val rs9061 Exon 5 C>T Glu>Lys rs1365776 Exon 8 A>G Gly>Arg rs3948464 Exon 11 G>A Leu>Ser rs1135791 Exon 14 T>C Met>Thr
relA v-rel reticuloendotheliosis viral 11q13 rs7101916 5'NUTR C>T oncogene homolog A rs11820062 Intron 1 C>T rs2306365 Intron 5 G>A rs11227247 Intron 10 T>G rs7119750 Intron 10 C>T Supplementary Table 2. Primers used for genotyping in this study SNP forward PCR primer (5′-3′) reverse PCR primer (5′-3′) extension primer (5′-3′)
MASSARRAY method rs1045845 ACGTTGGATGACCAGCCCCAACCCGC ACGTTGGATGAACCACCTTGATTGCA CCCCTGCGTCACCCACT TTT GCAG rs10852863 ACGTTGGATGTGCCCACTACAGGTTA ACGTTGGATGTGGACTGTTGCAAGTT GTGTACAACAGGAGATTGCC AATC CAGG rs3809849 ACGTTGGATGTTCGTGTTTCGGTGAG ACGTTGGATGACTCTGCGTCGCTTCTC CGTCGCTTCTCCAGGCGACATCGGCT TGTG CA rs7211078 ACGTTGGATGACAGAAGACCTAAAGC ACGTTGGATGAGGCTCAGTACCCTGA GCAGCCCCAATAAGAGCA AGCC TTTC rs751670 ACGTTGGATGACCCAAGTCGTGGCAG ACGTTGGATGAATGTTCTGACCAGCT CACCACCCACAGGCA TAG CTCG rs9907224 ACGTTGGATGGAACTGCACCAGGTGG ACGTTGGATGTCAGCACGCAGTTCAA GTGGTAGGTCCAGGG TAG GCAG rs9905742 ACGTTGGATGCCTTAGCCTTTGTCCTG ACGTTGGATGATGGCACACCTGCAGC TTCCTCTTCTTCCTGCCCA TTC CAC rs12603519 ACGTTGGATGGGCCCAGAGTTAAAAG ACGTTGGATGAAAGATGGGACGCCGC ACTTCCGCTCAGAGCTA AGCG TAGG rs11679983 ACGTTGGATGCTTCCCAAGTACCTCTA ACGTTGGATGTCCTTCAGTGCCCGCA CTTCAGTGCCCGCAATAGGCGGCT GTC ATAG rs11556887 ACGTTGGATGCAGAGACACACCAATC ACGTTGGATGTGGGGTATGGAGGGAG ATGGAGGGAGCTTCCTTCT CTAC CTT rs1365776 ACGTTGGATGAGATGTATCTGGTCAA ACGTTGGATGCTTCCTCTTGTACTCTC CCTCTTGTACTCTCATCTTACCTC CTCC ATC rs3948464 ACGTTGGATGCATGATGAGGGTCCAA ACGTTGGATGAGCAGATTCCATCATTA AAATGTGCCCGAAAGTCCAGAT AAGG GCC rs1135791 ACGTTGGATGGGAAGGAACGCAAAG ACGTTGGATGAATACCTTCAGCAGCT CTTCAGCAGCTCTCCTAGGGTC AACTG CTCC rs11820062 ACGTTGGATGAGCTGAATCAGGGCCT ACGTTGGATGTCAGTTTCCTCTCTGGA GGCCTGTTGTACTTTCTTAAGGAAAA GTTG GGG rs2306365 ACGTTGGATGGGGATATAGGTGCTATC ACGTTGGATGGGACTTGTGTTTCACA cCACACGCACCAGCGGCCCTCCCAG AG CACG rs7119750 ACGTTGGATGTCATGGTGCTCAGGCT ACGTTGGATGGGGTAAGAGGAAGGA GAGGAAGGAGATAGGGTGTT CAAT GATAG rs11227247 ACGTTGGATGGGGGAACAAAAATCAC ACGTTGGATGTGAAGTGCAGAGCTTG ATTAGCAAGATGTGTGTGCC CTC TCAG rs7101916 ACGTTGGATGAAGGCGCGTCCGTGTG ACGTTGGATGCCTGCAGTGGAGCATC CCCCACCCCAGGCCTCAT GAG CTC SNaPshot method rs11556887 CTGATGTGTGTCCCTTCACCA CCCTTCACAGTCACCTTGAGC CCCCCCCGTATGGAGGGAGCTTCCTTC T rs9061 CTGATGTGTGTCCCTTCACCA CCCTTCACAGTCACCTTGAGC CGGGCATCTCTTCTGAGTCTTCTT rs1135791 GCTGGCTTCAGTCCAAGAGAA TCCTAACGGGAGCTCTTCACA CCCCCCCCCCCCCCCTCAGCAGCTCTC CTAGGGTC Supplementary Table 3. The database for the meta-analysis included 5 studies on Sp110 polymorphisms with human pulmonary tuberculosis. The following items are listed for each study: first author, publication year, country and ethnicity of study subjects, characteristics of cases and controls, and method. First author Country Characteristics of cases Characteristics of controls Method (year) (ethnicity) Babb South Africa Patients had a microbiologically confirmed Controls were from a healthy population of the SNaPshot (2007) (Mixed race) diagnosis of pulmonary TB and were HIV- same ethnic and socioeconomic group as cases. negative. They had no history of TB. Szeszko Russia Patients’ diagnoses were confirmed by sputum Controls were local adult blood bank donors Taqman analysis (2007) (Russian) culture of MTB. Those with extra-pulmonary with no history of TB. method TB and/or HIV were excluded Cong China Patients were sputum smear-positive and Controls were from a healthy population PCR-RFLP plus (2010) (Chinese) confirmed by culture-positive for without pulmonary tuberculosis, other chronic sequencing mycobacteria and chest X-ray tuberculosis diseases or low immune function. Their levy like. They were without pneumonia, lung ethnicity was matched to cases’. cancer, pneumoconiosis with tuberculosis similar illness, HIV, EBV, or hepatitis. Liang China Patients were clinically and radiologically Controls were blood donors who were clinically SNaPshot (2011) (Chinese) diagnosed with pulmonary TB, and sputum normal. They had no history of TB, negative smears and cultures confirmed M. PPD tests, and no evidence of prior TB noted in tuberculosis infections. Patients were HIV- chest radiographies. There were matched to negative and none presented with other cases by region, ethnic background, age, and infectious diseases or with sex. immunosuppressive conditions Png Indonesia Patients were diagnosed with clinical Controls were non-genetically related to patients GoldenGate assay (2012) (Indonesian) symptoms, chest X-rays, and sputum smear, and lacked evidence of TB or TB-related plus Taqman and then diagnosis was confirmed by sputum infiltrates in chest X-rays. They were matched functionally tested culture of M. tuberculosis. Patients with extra- to cases by region of residence, age, sex, and assay. pulmonary TB, diabetes mellitus, and HIV ethnicity. were excluded Supplementary Table 4. Genotype frequencies of SNPs within Mybbp1a, SP110, and RelA genes in Chinese population SNP Genotypes Controls(%) Cases(%) OR(95% CI)* P p-trend† rs12603519 GG 221(52.00) 381(54.27) Ref GA 164(38.59) 270(38.46) 1.01[0.77-1.31] 0.97 0.26095 AA 40(9.41) 51(7.26) 0.84[0.67-1.05] 0.12 rs3809849 GG 301(70.82) 426(60.68) Ref GC 114(26.82) 239(34.05) 1.52[1.15-1.99] 0.003 0.00018 CC 10(2.36) 37(5.27) 1.55[1.08-2.23] 0.017 rs10852863 AA 116(27.29) 236(33.62) Ref AG 217(51.06) 341(48.58) 0.78[0.59-1.03] 0.078 0.01821 GG 92(21.65) 125(17.81) 0.80[0.67-0.95] 0.013 rs9907224 CC 101(23.76) 184(26.21) Ref CT 216(50.82) 341(48.58) 0.86[0.64-1.16] 0.332 0.54521 TT 108(25.41) 177(25.21) 0.94[0.79-1.11] 0.457 rs7211078 GG 98(23.11) 186(26.50) Ref GT 218(51.42) 314(44.73) 0.77[0.57-1.05] 0.097 0.98583 TT 108(25.47) 202(28.77) 0.99[0.84-1.18] 0.926 rs751670 TT 139(32.78) 246(35.04) Ref TC 216(50.94) 349(49.72) 0.91[0.70-1.19] 0.498 0.43202 CC 69(16.27) 107(15.24) 0.93[0.77-1.11] 0.420 rs9905742 TT 396(93.18) 618(88.03) Ref TA 28(6.59) 78(11.11) 1.74[1.11-2.73] 0.017 0.00426 AA 1(0.23) 6(0.86) 1.99[0.69-5.75] 0.204 rs1045845 CC 128(30.12) 237(33.91) Ref CG 209(49.18) 338(48.35) 0.88[0.67-1.16] 0.371 0.11853 GG 88(20.70) 124(17.74) 0.865[0.73-1.03] 0.105 rs11679983 GG 393(92.47) 661(94.16) Ref GA 31(7.29) 40(5.70) 0.77[0.47-1.25] 0.294 0.31658 AA 1(0.24) 1(0.14) 0.75[0.19-2.99] 0.679 rs11556887 CC 368(86.59) 566(80.63) Ref CT 54(12.71) 124(17.66) 1.60[1.12-2.28] 0.010 0.00709 TT 3(0.71) 12(1.71) 1.61[0.85-3.04] 0.142 rs9061 CC 277(65.18) 397(56.55) Ref CT 139(32.71) 268(38.18) 1.39[1.07-1.80] 0.013 8.30E-04 TT 9(2.11) 37(5.27) 1.72[1.18-2.51] 0.005 rs1365776 AA 357(84.00) 556(79.20) Ref AG 64(15.06) 132(18.80) 1.33[0.96-1.85] 0.090 0.03128 GG 4(0.94) 14(1.99) 1.47[0.84-2.59] 0.176 rs3948464 GG 402(94.59) 629(89.60) Ref GA 22(5.17) 72(10.26) 2.05[1.25-3.37] 0.004 0.00325 AA 1(0.24) 1(0.14) 0.80[0.20-3.19] 0.747 rs1135791 TT 281(66.12) 513(73.08) Ref TC 125(29.41) 176(25.07) 0.76[0.58-1.0] 0.048 0.003 CC 19(4.47) 13(1.85) 0.58[0.40-0.83] 0.003 rs7101916 CC 171(40.24) 290(41.31) Ref CT 200(47.06) 321(45.73) 0.96[0.74-1.24] 0.737 0.84424 TT 54(12.71) 91(12.96) 0.98[0.81-1.19] 0.835 rs11820062 CC 166(39.06) 244(34.76) Ref CT 186(43.76) 347(49.43) 1.28[0.98-1.67] 0.074 0.4935 TT 73(17.18) 111(15.81) 1.01 [0.84-1.21] 0.884 rs2306365 GG 160(37.65) 292(41.60) Ref GA 205(48.23) 303(43.16) 0.81[0.63-1.06] 0.124 0.50955 AA 60(14.12) 107(15.24) 0.96[0.79-1.16] 0.656 rs11227247 TT 160(37.65) 293(41.74) Ref TG 205(48.23) 302(43.02) 0.81[0.62-1.05] 0.113 0.48862 GG 60(14.12) 107(15.24) 0.96[0.79-1.15] 0.644 rs7119750 CC 160(37.65) 292(41.60) Ref CT 206(48.47) 303(43.16) 0.81[0.62-1.05] 0.116 0.5449 TT 59(13.88) 107(15.24) 0.97[0.80-1.17] 0.722 SNP indicates single nucleotide polymorphism; Ref,reference; OR,odds ration; CI, confidence interval * Adjusted for age and gender † P values were calculated by the Cochran-Armitage's trend test Supplementary Table 5. Results of meta–analysis for SP110 polymorphisms and TB 2 Contrast Comparisons(n) OR(95% CI)* P–value Ph I
rs1135791 T versus C alleles 5(7530) 1.11 [0.89–1.39] 0.36 0.007 72% (T/T+T/C) versus C/C 5(3765) 1.19 [0.73–1.92] 0.48 0.09 53% T/T versus (T/C+C/C) 5(3765) 1.12 [0.85–1.48] 0.41 0.004 74%
rs3948464 G versus A alleles 5(13612) 1.13 [0.82–1.55] 0.46 0.005 73% (G/G +A/G) versus A/A 5(6806) 0.92 [0.62–1.35] 0.67 0.45 0% G/G versus (A/G+ A/A) 5(6806) 1.05 [0.92–1.20] 0.46 0.002 76%
rs9061 C versus T alleles 4(5918) 1.24 [1.08–1.43] 0.002 0.13 47% (C/C+C/T) versus T/T 4(2959) 2.78 [1.16–6.66] 0.02 0.07 58% C/C versus (C/C+T/C) 4(2959) 1.18 [1.01–1.38] 0.04 0.16 42% OR (95% CI): odds ratio and 95% confidence interval. Ph: P values for heterogeneity for Q test.
* The fixed model was used when Ph >0.1 and the random model was used when Ph<0.1. Supplementary Figure 1. The SP110 SAND domain structure alteration caused by the residue mutant of T523M at the rs1135791 variant site. The 3D SAND structure with the Thr residue (A) is significantly different with the 3D SAND structure with the Met residue (B). The 3D structure of 1h5pa is described in (C). The structure is colored according to secondary structures: red, yellow and blue meanα-helix,β-sheet and turn.