Table 2. Supplement.
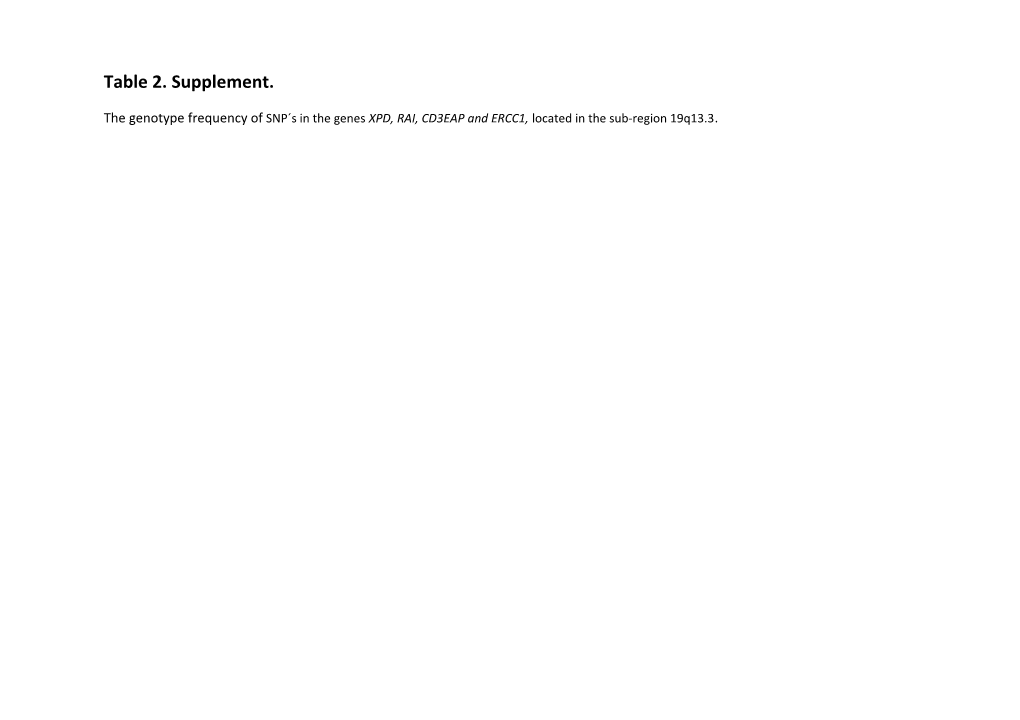
The genotype frequency of SNP´s in the genes XPD, RAI, CD3EAP and ERCC1, located in the sub-region 19q13.3. Trivial name systematic name Gene location Position Source of % Genotyped MAF HWE TTF OS
identification p value p value
XPD-exon23 *ERCC2 A>C 18123137 rs13181 96.5 C:0.388 0.02 0.003 0.05
XPD-exon10 *ERCC2 G>A 18135477 rs1799793 89.4 A:0.367 0.78 0.58 0.66
XPD-exon6 ERCC2 c.499A>C CGA-CGC 18136527 rs238406 67 T:0.466 0.92 0.12 0.45
XPD-intron5-1 ERCC2 c.492-62T>A Intron 18136696 rs238407 81 T:0.434 0.69 0.08 0.42
XPD-5´2 ERCC2 c.35-1958A>G Near gene3 18144005 rs2097215 70 G:0.402 0.74 0.25 0.52
RAI-3'9 NT_011109.15:g.18147886T>G Near PPP1R13L 3´ 18147886 rs10422489 73 G:0.387 0.81 0.57 0.82
før PPP1R13L Near PPP1R13L 3´ 18150199 rs4544343 73 G.0.391 0.53 0.77 RAI-3'4 NT_011109.15:g.18150199A>G 1.00
RAI-exon13-1 PPP1R13L c.*475T>A UTR-3 18151180 rs6966 80 T:0.151 0.30 0.15 0.65
RAI-intron8-1 *PPP1R13L c.1882-1425A>G Intron 18159091 rs1970764 94.5 G:0.185 0.16 0.88 0.17
RAI-intron8-12 PPP1R13L c.1881+756C>T Intron 18162599 rs12986272 83 A:0.253 0.68 0.39 0.69
RAI-intron1-7 PPP1R13L c.21-1931G>T Intron 18171746 rs10402584 73 T:0.496 0.78 0.47 0.48
RAI-intron1-10 PPP1R13L c.-21-2062G>A Intron 18171815 rs10402393 81 T:0.004 1.00 0.268 0.21
RAI-intron1-1 PPP1R13L c.21-2169A>G Intron 18171984 rs4572514 85.2 C: 0.175 0.88 0.003 0.02
RAI-intron1-4 PPP1R13L c.21-3291G>A Intron 18173106 rs4803816 84.6 T: 0.377 0.20 0.33 0.15
RAI-intron1-3 PPP1R13L c.22+734A>G Upstream 18175739 rs959457 85.5 C: 0.171 0.68 0.007 0.04
RAI-intron1-11 PPP1R13L c.-22+397C>G Upstream 18176078 rs928911 86.4 T: 0.052 1.00 0.05 0.30
RAI-intron1-5 PPP1R13L c.-22+289T>C Upstream 18176178 rs4803817 85.2 G: 0.206 0.44 0.95 0.40
RAI-intron1-12 NT_011109.15:g.18176179 18176179 Nr106 90.2 G: 0.215 0.74 0.80 0.77
RAI-5'1 NT_011109.15:g.18176679A>G Upstream 18176679 rs10412761 83.9 G: 0.375 0.17 0.25 0.15
ASE1-exon1 *CD3EAP c.468-21G>A Upstream 18178152 rs967591 96.8 A:0.177 0.27 0.002 0.004
ASE1-intron1-1 CD3EAP c.22+29A>C Intron 18178221 rs8113779 85.2 T: 0.167 0.83 0.007 0.02
ASE1-intron-2-1 CD3EAP Intron 18179548 rs2013521 82.6 T: 0.147 0.83 0.009 0.04
ASE1-exon3-1 CD3EAP c.1264A>G K261T 18180220 rs735482 84.6 C: 0.143 0.86 0.03 0.03
ASE1-exon3-2 CD3EAP c.1605A>G K375E 18180561 rs762562 85.8 G: 0.152 0.90 0.09 0.1
ASE1-exon3-3 CD3EAP c.1668G>A D396N 18180624 rs2336219 85.8 A: 0.115 1.00 0.03 0.05
ASE1-exon3-6 CD3EAP c.1998C>A Q506K 18180954 rs3212986 78.3 T: 0.254 0.69 0.02 0.03
ASE1-3'-1 CD3EAP 3`UTR 18181304 rs3212985 82.9 C: 0.205 0.10 0.77 0.45
ASE1-3'-2 CD3EAP 3´UTR 18181311 rs1007616 82.9 C: 0.379 0.11 0.78 0.66 * Previously published results (1). MAF: minor allele frequency
Reference List
(1) Vangsted A, Gimsing P, Klausen TW, Nexo BA, Wallin H, Andersen P, et al. Polymorphisms in the genes ERCC2, XRCC3 and CD3EAP influence treatment outcome in multiple myeloma patients undergoing autologous bone marrow transplantation. Int J Cancer 2007 Mar 1;120(5):1036- 45.