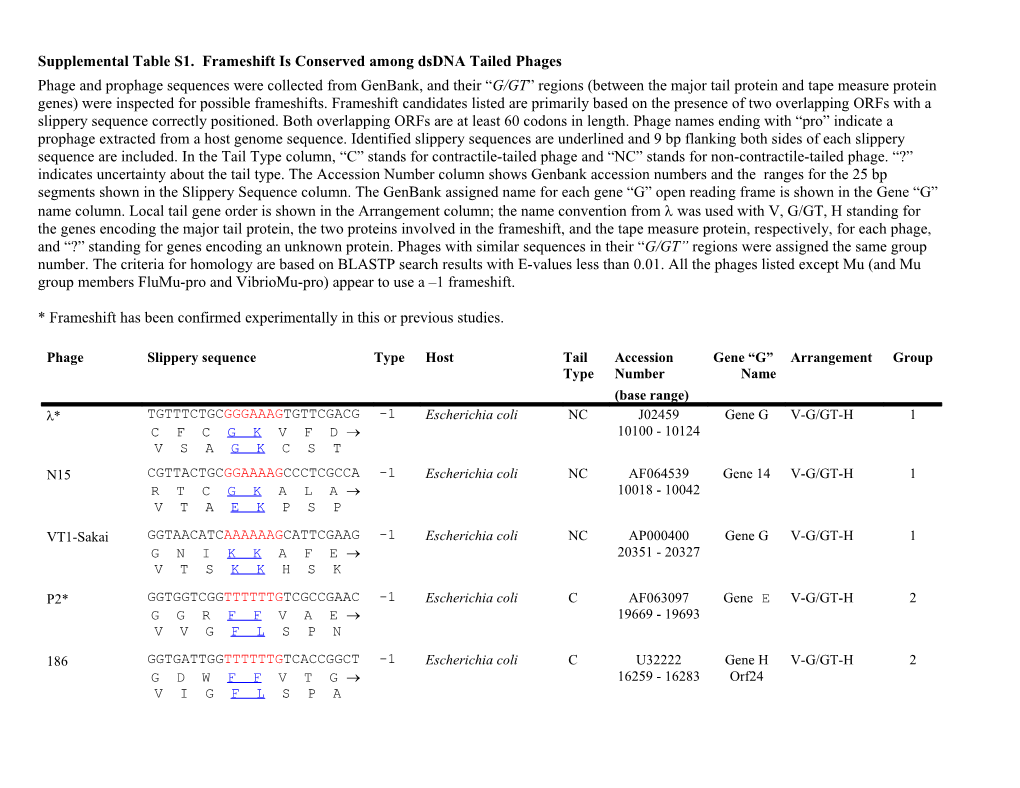
Supplemental Table S1. Frameshift Is Conserved among dsDNA Tailed Phages Phage and prophage sequences were collected from GenBank, and their “G/GT” regions (between the major tail protein and tape measure protein genes) were inspected for possible frameshifts. Frameshift candidates listed are primarily based on the presence of two overlapping ORFs with a slippery sequence correctly positioned. Both overlapping ORFs are at least 60 codons in length. Phage names ending with “pro” indicate a prophage extracted from a host genome sequence. Identified slippery sequences are underlined and 9 bp flanking both sides of each slippery sequence are included. In the Tail Type column, “C” stands for contractile-tailed phage and “NC” stands for non-contractile-tailed phage. “?” indicates uncertainty about the tail type. The Accession Number column shows Genbank accession numbers and the ranges for the 25 bp segments shown in the Slippery Sequence column. The GenBank assigned name for each gene “G” open reading frame is shown in the Gene “G” name column. Local tail gene order is shown in the Arrangement column; the name convention from was used with V, G/GT, H standing for the genes encoding the major tail protein, the two proteins involved in the frameshift, and the tape measure protein, respectively, for each phage, and “?” standing for genes encoding an unknown protein. Phages with similar sequences in their “G/GT” regions were assigned the same group number. The criteria for homology are based on BLASTP search results with E-values less than 0.01. All the phages listed except Mu (and Mu group members FluMu-pro and VibrioMu-pro) appear to use a –1 frameshift.
* Frameshift has been confirmed experimentally in this or previous studies.
Phage Slippery sequence Type Host Tail Accession Gene “G” Arrangement Group Type Number Name (base range) * TGTTTCTGCGGGAAAGTGTTCGACG -1 Escherichia coli NC J02459 Gene G V-G/GT-H 1 C F C G K V F D 10100 - 10124 V S A G K C S T N15 CGTTACTGCGGAAAAGCCCTCGCCA -1 Escherichia coli NC AF064539 Gene 14 V-G/GT-H 1 R T C G K A L A 10018 - 10042 V T A E K P S P
VT1-Sakai GGTAACATCAAAAAAGCATTCGAAG -1 Escherichia coli NC AP000400 Gene G V-G/GT-H 1 G N I K K A F E 20351 - 20327 V T S K K H S K
P2* GGTGGTCGGTTTTTTGTCGCCGAAC -1 Escherichia coli C AF063097 Gene E V-G/GT-H 2 G G R F F V A E 19669 - 19693 V V G F L S P N
186 GGTGATTGGTTTTTTGTCACCGGCT -1 Escherichia coli C U32222 Gene H V-G/GT-H 2 G D W F F V T G 16259 - 16283 Orf24 V I G F L S P A Fels-2 GGTGGTGACTTTTTTGTTGAAGAAG -1 Salmonella C AE008823 Gene V-G/GT-H 2 G G D F F V E E typhimurium 5268 - 5244 STM2699 V V T F L L K K
CTX GATCGCCGGTTTTTTGCTGCAGAAG -1 Pseudomonas C AB008550 orf24 V-G/GT-H 2 D R R F F A A E aeruginosa 19038 - 19062 I A G F L L Q K
L5* GACGCAACTGGGGGAAGCCGCGCCC -1 Mycobacterium NC Z18946 Gene 24 V-G/GT-H 3 D A T G G S R A 15349 - 15373 T Q L G E A A P
D29 GACGCAACTGGGGGAAGCCGCGCCC -1 Mycobacterium NC AF022214 gp24 V-G/GT-H 3 D A T G G S R A 16072 - 16096 T Q L G E A A P
Bxb1 GACGCAAGTGGGGGAAGCCTCCAGC -1 Mycobacterium NC AF271693 gp20 V-G/GT-H 3 D A S G G S L Q 15962 - 15986 T Q V G E A S S
Bxz2 GACCCAGCCGGGGGAAGCCGAGCGC -1 Mycobacterium NC AY129332 gp24 V-G/GT-H 3 D P A G G S R A 15639 - 15663 T Q P G E A E R
C31* GAACCTGAAGGGGAAGGCGAAGCCG -1 Streptomyces NC AJ006589 Gene 41 V-?-G/GT-H 4 E P E G E G E A 8639 – 8663 N L K G K A K P
TM4* GCTGATCGAGGGAAAATCTCGCAGG -1 Mycobacterium NC AF068845 gp15 V-G/GT-H 5 A D R G K I S Q 9724 – 9748 L I E G K S R R
Mu* ACACAAGAACGGGGGCGAGTGGCTG -2 Escherichia coli C NC_000929 Mup41 V-G/GT-H 6 H K N G G E W L 23948 - 23972 T Q E R G R V A
VibrioMu-pro CTTGAGGCTCGGGGGCGAGATTAAA -2 Vibrio cholerae C AE004256 Gene ?-G/GT-H 6 L R L G G E I K 3904 – 3880 VC1792 L E A R G R D *
FluMu-pro GTGGTCTCGCGGGGGCGATTGGATA -2 Haemophilus C U32827 Gene V-G/GT-H 6 W S R G G D W I influenzae 7930 – 7954 HI1513 V V S R G R L D HK97* CGAAGCGCGGGAAAAGTCTCAACCC -1 Escherichia coli NC AF069529 gp13 V-G/GT-M-H 7 R S A G K V S T 7825 – 7849 E A R E K S Q P
HK022* TAACGATCTGGGAAAGACTTCGAGC -1 Escherichia coli NC AF069308 Gene 13 V-G/GT-H 8 * R S G K D F E 8120 - 8144 N D L G K T S S
D3 GAACAACCTGGGAAAGACGACCAGC -1 Pseudomonas NC AF165214 Orf17 V-G/GT-H 8 E Q P G K D D Q aeruginosa 10928 - 10952 N N L G K T T S
SPP1 TAACTCAGGTTTTTTCAAACGAGCT -1 Bacillus subtilis NC X97918 Gene 17.5 V-?-G/GT-?-H 9 * L R F F Q T S 11680 - 11704 N S G F F K R A
PBSX AGAAGCAGCAAAAAACTAGTAAAAG -1 Bacillus subtilis C? Z99117 Gene yqbN ?-G/GT-H 10 R S S K K L V K 63233 -63209 E A A K N *
M2 AAATACTGAAAAAAAGAGTGGAACC -1 Methanobacterium NC AF065411 ORF19 V-G/GT-?-H 11 * K K E W N thermoautotrophicum 13703 - 13727 N T E K K S G T
M100 GGGTGTTAAGGGGAAGAAAAAGAAG -1 Methanothermobacter NC NC_002628 ORF19 V-G/GT-?-H 11 * G E E K E wolfei 20433 - 20457 G V K G K K K K
TP901-1 AAGCAATACGGGAAAGTTAATCAAA -1 Lactococcus NC AF304433 ORF43 V-G/GT-H 12 K Q T G K V N Q 22722 - 22746 S N T G K L I K r1t AAATAAAATAAAAAACTAATTCTCC -1 Lactococcus lactis NC? U38906 Orf38 V-G/GT- 13 * N K K L I L 22436 - 22460 H(split gene) N K I K N *
SM1 GAATCAGTTAAAAAAATAGCGCTCC -1 Streptococcus mitis NC AY007505 Gene 47 V-G/GT-H 13 E S V K K I A L 24339 - 24363 N Q L K K * c2 AGACGAAACAAAAAAAGCGATAGCA -1 Lactococcus NC L48605 Gene l8 V-G/GT-?-H 14 R R N K K S D S 12502 -12526 D E T K K A I A
BIL67 AGACGAAACAAAAAAAGCAATAGCA -1 Lactococcus NC L33769 Orf29 V-G/GT-?-H 14 R R N K K S D S 9178 - 9154 D E T K K A I A
ch1 CCGTGTTTTGGGAAATGCCGTCGAC -1 Natrialba magadii C NC_004084 ORF20 V-G/GT-?-H 15 P C F G K C R R 13530 - 13554 R V L G N A V D
105 TGAAGAGCCGGGGGAGTAAGTCTGT -1 Bacillus subtilis NC AB016282 ORF35 V-G/GT-H 16 * R A G G V S L 8892 – 8916 E E P G E *
PSA TTCAAAAGGAAAAAAATAAAACTGC -1 Listeria NC AJ312240 ORF11 V-G/GT-H 17 F K R K K I K L monocytogenes 7755 – 7779 S K G K K *
SfV TGTGGCTGGTTTTTTCCTCCAGGCC -1 Shigella flexneri C U82619 Orf13 V-G/GT-H 18 C G W F F P P G 9605 - 9629 V A G F F L Q A
-halo1-pro GACCGATAGTTTTTTCTACAAGAAG -1 Bacillus halodurans NC AP001510 Gene V-G/GT-H 19 * F F L Q E 131876 - BH0958 T D S F F Y K K 131900
-halo2-pro AGACCTGGGTTTTTTATTCCGTTAG -1 Bacillus halodurans NC AP001519 Gene V-G/GT-H 19 R P G F F I P L 38314 - 38290 BH3520 D L G F L F R *
F pyocin GGACGCGGAAAAAAACTGACCCCCG -1 Pseudomonas NC AB030825 PRF29 V-G/GT-H 20 G R G K K L T P aeruginosa 20677 – 20701 D A E K N *
PVL TAGCGATACAAAAAAGTTCGTGACA -1 Staphylococcus NC? AB009866 Orf 14 V-G/GT-H 21 * R T K K V R D aureus 8649 – 8673 S D T K K F V T
PV83 TAGCGATACAAAAAAGTTCGTGACA -1 Staphylococcus NC? AB044554 Orf 50 V-G/GT-H 21 * R Y K K V R D aureus 27323 - 27347 S D T K K F V T 01205 ATTTGATGATTTTTTATCAGCATTG -1 Streptococcus NC U88974 ORF38 V-G/GT-H 22 * F F I S I thermophilus 22598 - 22622 F D D F L S A L
Sfi11 ATTTGATGATTTTTTATCAGCATTG -1 Streptococcus NC AF158600 Orf117 V-G/GT-H 22 * F F I S I thermophilus 9758 - 9782 F D D F L S A L ul36 CGAAGAGTTAAAAAAGTCGGAATTT -1 Lactococcus lactiss NC AF349457 ORF116 V-G/GT-H 22 R R V K K V G I 25931 - 25955 E E L K K S E F
Sfi19 TGAAGCTGACCCAAAAGAATAAAAT -1 Streptococcus NC AF115102 Orf117 V-G/GT-H 23 * P K R I K thermophilus 9722 –9746 E A D P K E *
Sfi21 TGAAGTTGACCCAAAAGAATAAAGT -1 Streptococcus NC AF115103 Orf117 V-G/GT-H 23 * P K R I K thermophilus 8138 – 8162 E V D P K E *
7201 TGAAGCTGACCCAAAAGAATAAAAT -1 Streptococcus NC AF145054 orf32 V-G/GT-H 23 * P K R I K thermophilus 19466 - 19490 E A D P K E *
DT1 TGAAGTTGACCCAAAAGAATAAAAT -1 Streptococcus NC AF085222 Orf14 V-G/GT-H 23 * P K R I K thermophilus 8734 – 8758 E V D P K E *
L10 TGGCTCAACAAAAAAATAACAGCCC -1 Leuconostoc oenos NC L13035 OrfF V-G/GT-H 24 W L N K K I T A 2701 - 2725 G S T K K *
HP1 AATTACCGTAAAAAACTAACGGAGC -1 Haemophilus C U24159 Orf26 ?-G/GT-H 25 N T R K K L T E influenzae 21475 - 21499 I T V K N *
Yersini TGGTGAAGGGGCAAAGTAATTTCCC -1 Yersinia pestis NC AF074611 Gene V-G/GT-H 26 * R G K V I S 29665 - 29689 Y1042 G E G A K *
DeiPro CATTCCCGAGGGAAAAGCCCTGGAG -1 Deinococcus C? AE001862 DRA0106 ?-G/GT-H 27 H S R G K S P G radiodurans 99031 - 99055 I P E G K A L E Borreliapro GCGTGTTTCAAAAAAGTAGAGCTTG -1 Borrelia ? AE000786 BBG12 ?-G/GT-H 28 A C F K K V E L burgdorferi 11081 - 11057 R V S K K *