Brownian Dynamics Simulations of Interaction Between Scorpion Toxin Lq2 and Potassium Ion Channel
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Dynamics Simulation Reveals Specific Interaction
toxins Article Molecular Dynamics Simulation Reveals Specific Interaction Sites between Scorpion Toxins and Kv1.2 Channel: Implications for Design of Highly Selective Drugs Shouli Yuan 1,2, Bin Gao 1 and Shunyi Zhu 1,* ID 1 Group of Peptide Biology and Evolution, State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; [email protected] (S.Y.); [email protected] (B.G.) 2 College of Resources and Environment, University of Chinese Academy of Sciences, Beijing 100049, China * Correspondence: [email protected] Academic Editors: Bryan Grieg Fry and Steve Peigneur Received: 29 August 2017; Accepted: 19 October 2017; Published: 1 November 2017 Abstract: The Kv1.2 channel plays an important role in the maintenance of resting membrane potential and the regulation of the cellular excitability of neurons, whose silencing or mutations can elicit neuropathic pain or neurological diseases (e.g., epilepsy and ataxia). Scorpion venom contains a variety of peptide toxins targeting the pore region of this channel. Despite a large amount of structural and functional data currently available, their detailed interaction modes are poorly understood. In this work, we choose four Kv1.2-targeted scorpion toxins (Margatoxin, Agitoxin-2, OsK-1, and Mesomartoxin) to construct their complexes with Kv1.2 based on the experimental structure of ChTx-Kv1.2. Molecular dynamics simulation of these complexes lead to the identification of hydrophobic patches, hydrogen-bonds, and salt bridges as three essential forces mediating the interactions between this channel and the toxins, in which four Kv1.2-specific interacting amino acids (D353, Q358, V381, and T383) are identified for the first time. -

X-Ray Fluorescence Analysis Method Röntgenfluoreszenz-Analyseverfahren Procédé D’Analyse Par Rayons X Fluorescents
(19) & (11) EP 2 084 519 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: G01N 23/223 (2006.01) G01T 1/36 (2006.01) 01.08.2012 Bulletin 2012/31 C12Q 1/00 (2006.01) (21) Application number: 07874491.9 (86) International application number: PCT/US2007/021888 (22) Date of filing: 10.10.2007 (87) International publication number: WO 2008/127291 (23.10.2008 Gazette 2008/43) (54) X-RAY FLUORESCENCE ANALYSIS METHOD RÖNTGENFLUORESZENZ-ANALYSEVERFAHREN PROCÉDÉ D’ANALYSE PAR RAYONS X FLUORESCENTS (84) Designated Contracting States: • BURRELL, Anthony, K. AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Los Alamos, NM 87544 (US) HU IE IS IT LI LT LU LV MC MT NL PL PT RO SE SI SK TR (74) Representative: Albrecht, Thomas Kraus & Weisert (30) Priority: 10.10.2006 US 850594 P Patent- und Rechtsanwälte Thomas-Wimmer-Ring 15 (43) Date of publication of application: 80539 München (DE) 05.08.2009 Bulletin 2009/32 (56) References cited: (60) Divisional application: JP-A- 2001 289 802 US-A1- 2003 027 129 12164870.3 US-A1- 2003 027 129 US-A1- 2004 004 183 US-A1- 2004 017 884 US-A1- 2004 017 884 (73) Proprietors: US-A1- 2004 093 526 US-A1- 2004 235 059 • Los Alamos National Security, LLC US-A1- 2004 235 059 US-A1- 2005 011 818 Los Alamos, NM 87545 (US) US-A1- 2005 011 818 US-B1- 6 329 209 • Caldera Pharmaceuticals, INC. US-B2- 6 719 147 Los Alamos, NM 87544 (US) • GOLDIN E M ET AL: "Quantitation of antibody (72) Inventors: binding to cell surface antigens by X-ray • BIRNBAUM, Eva, R. -

Modeling of the Binding of Peptide Blockers to Voltage-Gated Potassium Channels: Approaches and Evidence
REVIEWS Modeling of the Binding of Peptide Blockers to Voltage-Gated Potassium Channels: Approaches and Evidence V. N. Novoseletsky1*, A. D. Volyntseva1, K. V. Shaitan1, M. P. Kirpichnikov1,2, A. V. Feofanov1,2 1M.V.Lomonosov Moscow State University, Faculty of Biology, Leninskie Gory 1, bldg. 12, 119992, Moscow, Russia 2Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Miklukho- Maklaya str. 16/10, 117997, Moscow, Russia *Email: [email protected] Received: 06.07.2015 Copyright © 2016 Park-media, Ltd. This is an open access article distributed under the Creative Commons Attribution License,which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Modeling of the structure of voltage-gated potassium (KV) channels bound to peptide blockers aims to identify the key amino acid residues dictating affinity and provide insights into the toxin-channel interface. Computational approaches open up possibilities for in silico rational design of selective blockers, new molecular tools to study the cellular distribution and functional roles of potassium channels. It is anticipated that optimized blockers will advance the development of drugs that reduce over activation of potassium channels and attenuate the associated malfunction. Starting with an overview of the recent advances in computational simulation strat- egies to predict the bound state orientations of peptide pore blockers relative to KV-channels, we go on to review algorithms -

A Four-Disulphide-Bridged Toxin, with High Affinity Towards Voltage-Gated
Biochem. J. (1997) 328, 321–327 (Printed in Great Britain) 321 A four-disulphide-bridged toxin, with high affinity towards voltage-gated K+ channels, isolated from Heterometrus spinnifer (Scorpionidae) venom ! Bruno LEBRUN*1,Regine ROMI-LEBRUN*, Marie-France MARTIN-EAUCLAIRE†, Akikazu YASUDA*, Masaji ISHIGURO*, Yoshiaki OYAMA‡, Olaf PONGS§ and Terumi NAKAJIMA* *Suntory Institute for Bioorganic Research, Mishima-Gun, Shimamoto-Cho, Wakayamadai 1-1-1, 618 Osaka, Japan, †Laboratoire de Biochimie, CNRS UMR 6560, Faculte! de Me! decine Nord, 13916 Marseille Cedex 20, France, ‡Suntory Ltd Institute for Biomedical Research, Mishima-Gun, Shimamoto-Cho, Wakayamadai 1-1-1, 618 Osaka, Japan, and §Zentrum fu$ r Molekulare Neurobiologie, Institute fu$ r Neurale Signalverarbeitung, D-20246 Hamburg, Federal Republic of Germany A new toxin, named HsTX1, has been identified in the venom of limited reduction–alkylation at acidic pH and (2) enzymic Heterometrus spinnifer (Scorpionidae), on the basis of its ability cleavage on an immobilized trypsin cartridge, both followed by to block the rat Kv1.3 channels expressed in Xenopus oocytes. mass and sequence analyses. Three of the disulphide bonds are HsTX1 has been purified and characterized as a 34-residue connected as in the three-disulphide-bridged scorpion toxins, peptide reticulated by four disulphide bridges. HsTX1 shares and the two extra half-cystine residues of HsTX1 are cross- 53% and 59% sequence identity with Pandinus imperator toxin1 linked, as in Pi1. These results, together with those of CD (Pi1) and maurotoxin, two recently isolated four-disulphide- analysis, suggest that HsTX1 probably adopts the same general bridged toxins, whereas it is only 32–47% identical with the folding as all scorpion K+ channel toxins. -

Venom‑Derived Peptide Modulators of Cation‑Selective Channels : Friend, Foe Or Frenemy
This document is downloaded from DR‑NTU (https://dr.ntu.edu.sg) Nanyang Technological University, Singapore. Venom‑derived peptide modulators of cation‑selective channels : friend, foe or frenemy Bajaj, Saumya; Han, Jingyao 2019 Bajaj, S., & Han, J. (2019). Venom‑Derived Peptide Modulators of Cation‑Selective Channels: Friend, Foe or Frenemy. Frontiers in Pharmacology, 10, 58‑. doi:10.3389/fphar.2019.00058 https://hdl.handle.net/10356/88522 https://doi.org/10.3389/fphar.2019.00058 © 2019 Bajaj and Han. This is an open‑access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms. Downloaded on 30 Sep 2021 04:20:36 SGT fphar-10-00058 February 23, 2019 Time: 18:29 # 1 MINI REVIEW published: 26 February 2019 doi: 10.3389/fphar.2019.00058 Venom-Derived Peptide Modulators of Cation-Selective Channels: Friend, Foe or Frenemy Saumya Bajaj*† and Jingyao Han† Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore Ion channels play a key role in our body to regulate homeostasis and conduct electrical signals. With the help of advances in structural biology, as well as the discovery of numerous channel modulators derived from animal toxins, we are moving toward a better understanding of the function and mode of action of ion channels. -

Towards Therapeutic Applications of Arthropod Venom K+-Channel Blockers in CNS Neurologic Diseases Involving Memory Acquisition and Storage
Hindawi Publishing Corporation Journal of Toxicology Volume 2012, Article ID 756358, 21 pages doi:10.1155/2012/756358 Review Article Towards Therapeutic Applications of Arthropod Venom K+-Channel Blockers in CNS Neurologic Diseases Involving Memory Acquisition and Storage Christiano D. C. Gati,1, 2 Marcia´ R. Mortari,1 and Elisabeth F. Schwartz1 1 Departamento de Ciˆencias Fisiologicas,´ Instituto de Ciˆencias Biologicas,´ Universidade de Bras´ılia, 70910-900 Bras´ılia, DF, Brazil 2 Universidade Catolica´ de Bras´ılia, 71966-700 Bras´ılia, DF, Brazil Correspondence should be addressed to Elisabeth F. Schwartz, [email protected] Received 29 December 2011; Accepted 8 February 2012 Academic Editor: Yonghua Ji Copyright © 2012 Christiano D. C. Gati et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Potassium channels are the most heterogeneous and widely distributed group of ion channels and play important functions in all cells, in both normal and pathological mechanisms, including learning and memory processes. Being fundamental for many diverse physiological processes, K+-channels are recognized as potential therapeutic targets in the treatment of several Central Nervous System (CNS) diseases, such as multiple sclerosis, Parkinson’s and Alzheimer’s diseases, schizophrenia, HIV-1-associated dementia, and epilepsy. Blockers of these channels are therefore potential candidates for the symptomatic treatment of these neuropathies, through their neurological effects. Venomous animals have evolved a wide set of toxins for prey capture and defense. These compounds, mainly peptides, act on various pharmacological targets, making them an innumerable source of ligands for answering experimental paradigms, as well as for therapeutic application. -

Potassium Channels: from Scorpion Venoms to High-Resolution Structure
Toxicon 39 (2001) 739±748 Review www.elsevier.com/locate/toxicon Potassium channels: from scorpion venoms to high-resolution structure M.L. Garciaa,*, Ying-Duo Gaob, O.B. McManusa, G.J. Kaczorowskia aDepartment of Membrane Biochemistry and Biophysics, Merck Research Laboratories, P.O. Box 2000, Rahway, NJ 07065, USA bDepartment of Molecular Systems, Merck Research Laboratories, P.O. Box 2000, Rahway, NJ 07065, USA Received 8 June 2000; accepted 5 July 2000 1. Introduction the efforts of the many research laboratories that have focused on the study of K1 channels: (1) the extensive clon- In 1998, the ®eld of ion channel research entered a new ing and functional expression of these proteins; and (2) the era when the ®rst, high-resolution crystal structure of one of existence of a large number of high af®nity peptidyl inhibi- these proteins was solved (Doyle et al., 1998). For the ®rst tors of these proteins, isolated from different scorpion and time, it was possible to understand, at a molecular level, the spider venoms (Tytgat et al., 1999). In fact, the use of pep- mechanisms that control ion selectivity and conduction in tidyl inhibitors derived from scorpion venoms provided the potassium channels. The protein whose structure had been ®rst indirect information concerning K1 channel structure. determined, the KcsA K1 channel, is a two transmembrane For instance, both identi®cation of the pore region of the spanning domain, potassium selective channel from Strepto- channel (MacKinnon and Miller, 1989b), and determination myces lividans that gates in response to H1 when reconsti- of the tetrameric composition of K1 channels (MacKinnon, tuted in arti®cial lipid bilayers (Cuello et al., 1998; 1991) were made possible with the use of scorpion toxins. -
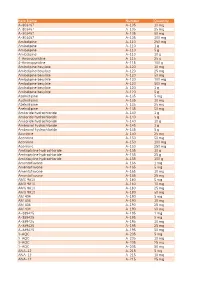
Item Name Catalog Number Quantity A-803467 A-105 10 Mg A
Catalog Item Name Number Quantity A-803467 A-105 10 mg A-803467 A-105 25 mg A-803467 A-105 50 mg A-803467 A-105 100 mg Amlodipine A-110 250 mg Amlodipine A-110 1 g Amlodipine A-110 5 g Amlodipine A-110 10 g 4-Aminopyridine A-115 25 g 4-Aminopyridine A-115 100 g Amlodipine besylate A-120 10 mg Amlodipine besylate A-120 25 mg Amlodipine besylate A-120 50 mg Amlodipine besylate A-120 100 mg Amlodipine besylate A-120 500 mg Amlodipine besylate A-120 1 g Amlodipine besylate A-120 5 g Azelnidipine A-135 5 mg Azelnidipine A-135 10 mg Azelnidipine A-135 25 mg Azelnidipine A-135 50 mg Amiloride hydrochloride A-140 1 g Amiloride hydrochloride A-140 5 g Amiloride hydrochloride A-140 10 g Ambroxol hydrochloride A-145 1 g Ambroxol hydrochloride A-145 5 g Aconitine A-150 25 mg Aconitine A-150 50 mg Aconitine A-150 100 mg Aconitine A-150 250 mg Amitriptyline hydrochloride A-155 10 g Amitriptyline hydrochloride A-155 25 g Amitriptyline hydrochloride A-155 100 g Amentoflavone A-165 1 mg Amentoflavone A-165 5 mg Amentoflavone A-165 10 mg Amentoflavone A-165 25 mg AMG 9810 A-180 5 mg AMG 9810 A-180 10 mg AMG 9810 A-180 25 mg AMG 9810 A-180 50 mg AM 404 A-190 5 mg AM 404 A-190 10 mg AM 404 A-190 25 mg AM 404 A-190 50 mg A-889425 A-195 1 mg A-889425 A-195 5 mg A-889425 A-195 10 mg A-889425 A-195 25 mg A-889425 A-195 50 mg 3-AQC A-205 5 mg 3-AQC A-205 10 mg 3-AQC A-205 25 mg 3-AQC A-205 50 mg ANA-12 A-215 5 mg ANA-12 A-215 10 mg ANA-12 A-215 25 mg ANA-12 A-215 50 mg ANA-12 A-215 100 mg A 967079 A-225 5 mg A 967079 A-225 10 mg A 967079 A-225 25 mg A 967079 -

Modeling the Structure of Agitoxin in Complex With
Biophysical Journal Volume 83 November 2002 2595–2609 2595 Modeling the Structure of Agitoxin in Complex with the Shaker K؉ Channel: A Computational Approach Based on Experimental Distance Restraints Extracted from Thermodynamic Mutant Cycles Mats A. L. Eriksson and Benoıˆt Roux Weill Medical College of Cornell University, Department of Biochemistry, New York, New York 10021 USA ABSTRACT Computational methods are used to determine the three-dimensional structure of the Agitoxin (AgTx2) -Shaker complex. In a first stage, a large number of models of the complex are generated using high temperature molecular dynamics, accounting for side chain flexibility with distance restraints deduced from thermodynamic analysis of double mutant cycles. Four plausible binding mode candidates are found using this procedure. In a second stage, the quality and validity of the resulting complexes is assessed by examining the stability of the binding modes during molecular dynamics simulations with explicit water molecules and by calculating the binding free energies of mutant proteins using a continuum solvent representation and comparing with experimental data. The docking protocol and the continuum solvent model are validated using the Barstar-Barnase and the lysozyme-antibody D1.2 complexes, for which there are high-resolution structures as well as double mutant data. This combination of computational methods permits the identification of two possible structural models of AgTx2 in complex with the Shaker Kϩ channel, additional structural analysis providing further evidence in favor of a single model. In this final complex, the toxin is bound to the extracellular entrance of the channel along the pore axis via a combination of hydrophobic, hydrogen bonding, and electrostatic interactions. -

Structural and Functional Study of Potassium Channel Inhibitor Hstx1
Structural and functional study of potassium channel inhibitor HsTX1 Mao-Feng Ger Introduction Membrane proteins are thought to account for 30% of genes; thus there may be at least 10 000 membrane proteins encoded in the human genome. Ion channels are integral membrane proteins that allow movement of ions across membranes down their electro- chemical gradients. These channel proteins form water-filled, gated pores that are often highly selective for specific ions (such as sodium, calcium, potassium or chloride). Only when the channels are open, ions can flow in and out. The opening of ion channels depends on of membrane potential (voltage-gated channels), binding of signaling molecules such as neurotransmitters, ions or nucleotides (ligand-gated channels) or stretch of the membrane (mechanosensitive channels). Potassium channels comprise a large family of ion channels. Their physiological functions are quite important, concerned with maintaining a negative voltage inside cells relative to outside. K channels have different families, according to their gating mechanism, i.e. the control of opening and closing of the channel. All K channels share the same core structure. Voltage-gated potassium channel is made from 4 alpha polypeptides forming a central pore. Each polypeptide has 6 transmembrane regions. The beta chain is regulatory and can interact with the alpha subunit to regulate the gating kinetics and enhance the stability of the complex. Another class of potassium channels is in-ward rectifiers. The channel is made from 4 identical subunits. They have 2 membrane-spanning segments and 1 pore-lining segment. Analysis of K channel sequences can understand the relationships between sequence motifs and various aspects of physiological function. -

Ion Channel-Target Toxicology
Journal of Toxicology Ion Channel-Target Toxicology Guest Editors: Yonghua Ji, Jan Tytgat, Maria Elena de Lima, Hongzhuan Chen, and Yun Zhang Ion Channel-Target Toxicology Journal of Toxicology Ion Channel-Target Toxicology Guest Editors: Yonghua Ji, Jan Tytgat, Maria Elena de Lima, Hongzhuan Chen, and Yun Zhang Copyright © 2012 Hindawi Publishing Corporation. All rights reserved. This is a special issue published in “Journal of Toxicology.” All articles are open access articles distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is prop- erly cited. Editorial Board Syed F. Ali, USA Hisato Iwata, Japan Gary H. Perdew, USA Michael Aschner, USA Margaret James, USA Cinta Porte, Spain Thomas Burbacher, USA Yujian James Kang, USA Robert H. Rice, USA Steven J. Bursian, USA Mary Kanz, USA Rudy Richardson, USA James Bus, USA M. Firoze Khan, USA Arleen Rifkind, USA Lucio Guido Costa, USA Paul Kostyniak, USA JeanClare Seagrave, USA Edmond Edmond Creppy, France Robert Krieger, USA James Sikarskie, USA Kevin Crofton, USA Kannan Krishnan, Canada J. J. Stegeman, USA Michael L. Cunningham, USA B. L. Lasley, USA Susan Sumner, USA Anthony DeCaprio, USA Pamela Lein, USA Robert Tanguay, USA David Doolittle, USA Robert Luebke, USA Kenneth Turteltaub, USA Paul R. Ebert, Australia Michael R. Moore, Australia Brad Upham, USA Laurence D. Fechter, USA Jack Ng, Australia William Valentine, USA M. Teresa Colomina Fosch, Spain P. J. O’Brien, Canada J. -

Small Gtpases* 1,2 3§ David J
Small GTPases* 1,2§ 3§ David J. Reiner and Erik A. Lundquist 1Institute of Biosciences and Technology, Texas A&M Health Science Center, Houston, TX USA 2Department of Medical Physiology, College of Medicine, Texas A&M University Health Science Center, Temple, TX USA 3Department of Molecular Biosciences, University of Kansas, Lawrence, KS USA Table of Contents 1. Overview ...............................................................................................................................3 2. The Ras family ........................................................................................................................7 2.1. LET-60 .......................................................................................................................7 2.2. RAS-1 .........................................................................................................................9 2.3. RAS-2 ....................................................................................................................... 10 2.4. RAP-1 ....................................................................................................................... 10 2.5. RAP-2 ....................................................................................................................... 11 2.6. RAP-3 ....................................................................................................................... 11 2.7. RAL-1 .....................................................................................................................